6S90
 
 | | BTK in complex with an inhibitor | | Descriptor: | 4-~{tert}-butyl-~{N}-[2-methyl-3-[6-[4-(4-methylpiperazin-1-yl)carbonylphenyl]-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]phenyl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Gutmann, S, Hinniger, A. | | Deposit date: | 2019-07-11 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Design of Potent and Selective Covalent Inhibitors of Bruton's Tyrosine Kinase Targeting an Inactive Conformation.
Acs Med.Chem.Lett., 10, 2019
|
|
8CPO
 
 | | Crystal structure of the PolB16_OarG intein variant S1A, N183A, C111A, C165A | | Descriptor: | PolB16 Intein Cys-less | | Authors: | Kattelmann, S, Pasch, T, Mootz, H.D, Kuemmel, D. | | Deposit date: | 2023-03-03 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analysis of a novel atypically split intein reveals a conserved histidine specific to cysteine-less inteins.
Chem Sci, 14, 2023
|
|
8CPN
 
 | | Crystal structure of the PolB16_OarG intein variant S1A, N183A | | Descriptor: | IODIDE ION, PolB16 intein | | Authors: | Kattelmann, S, Pasch, T, Mootz, H.D, Kummel, D. | | Deposit date: | 2023-03-03 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical analysis of a novel atypically split intein reveals a conserved histidine specific to cysteine-less inteins.
Chem Sci, 14, 2023
|
|
5O83
 
 | | Discovery of CDZ173 (leniolisib), Representing a Structurally Novel Class of PI3K Delta-Selective Inhibitors | | Descriptor: | Leniolisib, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Gutmann, S, Rummel, G, Shrestha, B. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of CDZ173 (Leniolisib), Representing a Structurally Novel Class of PI3K Delta-Selective Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
4BPG
 
 | | Crystal structure of Bacillus subtilis DltC | | Descriptor: | D-ALANINE--POLY(PHOSPHORIBITOL) LIGASE SUBUNIT 2 | | Authors: | Yonus, H, Zimmermann, S, Neumann, P, Stubbs, M.T. | | Deposit date: | 2013-05-26 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-Resolution Structures of the D-Alanyl Carrier Protein (Dcp) Dltc from Bacillus Subtilis Reveal Equivalent Conformations of Apo- and Holo-Forms
FEBS Lett., 589, 2015
|
|
7R26
 
 | | PI3K delta in complex with SD5 | | Descriptor: | 5-[2,6-di(morpholin-4-yl)pyrimidin-4-yl]-4-(trifluoromethyl)pyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Gutmann, S, Rummel, G, Shrestha, B. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor.
J.Med.Chem., 65, 2022
|
|
7R2B
 
 | | PI3Kdelta in complex with an inhibitor | | Descriptor: | (4~{S})-3-[6-[2-azanyl-4-(trifluoromethyl)pyrimidin-5-yl]-2-morpholin-4-yl-pyrimidin-4-yl]-4-methyl-1,3-oxazolidin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Gutmann, S, Rummel, G, Shrestha, B. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor.
J.Med.Chem., 65, 2022
|
|
6NTD
 
 | | Crystal Structure of G12V HRas-GppNHp bound in complex with the engineered RBD variant 12 of CRAF Kinase protein | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Maisonneuve, P, Kurinov, I, Wiechmann, S, Ernst, A, Sicheri, F. | | Deposit date: | 2019-01-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Conformation-specific inhibitors of activated Ras GTPases reveal limited Ras dependency of patient-derived cancer organoids.
J.Biol.Chem., 295, 2020
|
|
6NTC
 
 | | Crystal Structure of G12V HRas-GppNHp bound in complex with the engineered RBD variant 1 of CRAF Kinase protein | | Descriptor: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Maisonneuve, P, Kurinov, I, Wiechmann, S, Ernst, A, Sicheri, F. | | Deposit date: | 2019-01-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conformation-specific inhibitors of activated Ras GTPases reveal limited Ras dependency of patient-derived cancer organoids.
J.Biol.Chem., 295, 2020
|
|
5IS5
 
 | |
5IU2
 
 | | Discovery of imidazoquinolines as a novel class of potent, selective and in vivo efficacious COT kinase inhibitors | | Descriptor: | Mitogen-activated protein kinase kinase kinase 8, N-[2-(morpholin-4-yl)ethyl]-6-(8-phenyl-1H-imidazo[4,5-c][1,7]naphthyridin-1-yl)-1,3-benzothiazol-2-amine | | Authors: | Gutmann, S, Hinniger, A. | | Deposit date: | 2016-03-17 | | Release date: | 2016-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Imidazoquinolines as a Novel Class of Potent, Selective, and in Vivo Efficacious Cancer Osaka Thyroid (COT) Kinase Inhibitors.
J.Med.Chem., 59, 2016
|
|
1MLZ
 
 | | Crystal Structure of 7,8-Diaminopelargonic Acid Synthase in complex with the trans-isomer of amiclenomycin. | | Descriptor: | 7,8-diamino-pelargonic acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sandmark, J, Mann, S, Marquet, A, Schneider, G. | | Deposit date: | 2002-09-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the inhibition of the biosynthesis of biotin by the antibiotic amiclenomycin
J.Biol.Chem., 277, 2002
|
|
1MLY
 
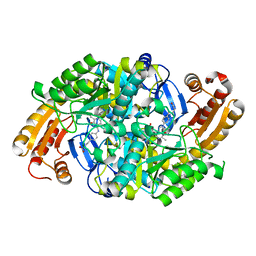 | | Crystal Structure of 7,8-Diaminopelargonic Acid Synthase in complex with the cis isomer of amiclenomycin | | Descriptor: | 7,8-diamino-pelargonic acid aminotransferase, CIS-AMICLENOMYCIN, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Sandmark, J, Mann, S, Marquet, A, Schneider, G. | | Deposit date: | 2002-09-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for the inhibition of the biosynthesis of biotin by the antibiotic amiclenomycin
J.Biol.Chem., 277, 2002
|
|
1QRQ
 
 | |
4V94
 
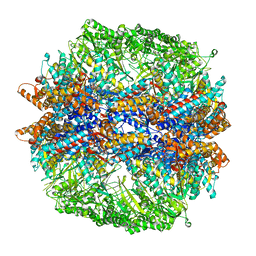 | | Molecular architecture of the eukaryotic chaperonin TRiC/CCT derived by a combination of chemical crosslinking and mass-spectrometry, XL-MS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Leitner, A, Joachimiak, L.A, Bracher, A, Walzthoeni, T, Chen, B, Monkemeyer, L, Pechmann, S, Holmes, S, Cong, Y, Ma, B, Ludtke, S, Chiu, W, Hartl, F.U, Aebersold, R, Frydman, J. | | Deposit date: | 2012-01-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Architecture of the Eukaryotic Chaperonin TRiC/CCT.
Structure, 20, 2012
|
|
6OY4
 
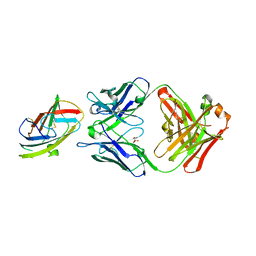 | | Crystal structure of complex between recombinant Der p 2.0103 and Fab fragment of 7A1 | | Descriptor: | Der p 2 variant 3, Fab fragment of IgG, HEAVY CHAIN, ... | | Authors: | Kapingidza, A.B, Offermann, L.R, Glesner, J, Wunschmann, S, Vailes, L.D, Chapman, M.D.C, Pomes, A, Chruszcz, M. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Human IgE Antibody Binding Site on Der p 2 for the Design of a Recombinant Allergen for Immunotherapy.
J Immunol., 203, 2019
|
|
1M22
 
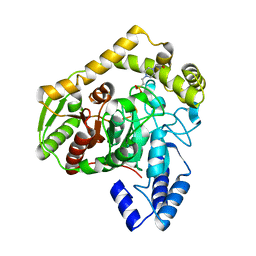 | | X-ray structure of native peptide amidase from Stenotrophomonas maltophilia at 1.4 A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, peptide amidase | | Authors: | Labahn, J, Neumann, S, Buldt, G, Kula, M.-R, Granzin, J. | | Deposit date: | 2002-06-21 | | Release date: | 2002-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An alternative mechanism for amidase signature enzymes
J.MOL.BIOL., 322, 2002
|
|
1M21
 
 | | Crystal structure analysis of the peptide amidase PAM in complex with the competitive inhibitor chymostatin | | Descriptor: | CHYMOSTATIN, Peptide Amidase | | Authors: | Labahn, J, Neumann, S, Buldt, G, Kula, M.-R, Granzin, J. | | Deposit date: | 2002-06-21 | | Release date: | 2002-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An alternative mechanism for amidase signature enzymes
J.MOL.BIOL., 322, 2002
|
|
4IRN
 
 | | Crystal Structure of the Prolyl Acyl Carrier Protein Oxidase AnaB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Prolyl-ACP dehydrogenase | | Authors: | Moncoq, K, Mann, S, Regad, L, Mejean, A, Ploux, O. | | Deposit date: | 2013-01-15 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the prolyl-acyl carrier protein oxidase involved in the biosynthesis of the cyanotoxin anatoxin-a.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4WWD
 
 | | High-resolution structure of the Co-bound form of the Y135F mutant of C. metallidurans CnrXs | | Descriptor: | CARBON DIOXIDE, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Volbeda, A, Coves, J, Maillard, A.P, Kuennemann, S, Grosse, C, Schleuder, G, Petit-Haertlein, I, de Rosny, E, Nies, D.H. | | Deposit date: | 2014-11-10 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Response of CnrX from Cupriavidus metallidurans CH34 to nickel binding.
Metallomics, 7, 2015
|
|
4WWB
 
 | | High-resolution structure of the Ni-bound form of the Y135F mutant of C. metallidurans CnrXs | | Descriptor: | CARBON DIOXIDE, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Volbeda, A, Coves, J, Maillard, A.P, Kuennemann, S, Grosse, C, Schleuder, G, Petit-Haertlein, I, de Rosny, E, Nies, D.H. | | Deposit date: | 2014-11-10 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Response of CnrX from Cupriavidus metallidurans CH34 to nickel binding.
Metallomics, 7, 2015
|
|
1EXB
 
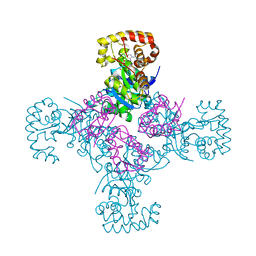 | | STRUCTURE OF THE CYTOPLASMIC BETA SUBUNIT-T1 ASSEMBLY OF VOLTAGE-DEPENDENT K CHANNELS | | Descriptor: | KV BETA2 PROTEIN, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM CHANNEL KV1.1 | | Authors: | Gulbis, J.M, Zhou, M, Mann, S, MacKinnon, R. | | Deposit date: | 2000-05-02 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the cytoplasmic beta subunit-T1 assembly of voltage-dependent K+ channels.
Science, 289, 2000
|
|
2G32
 
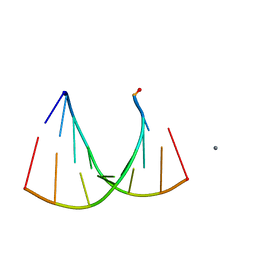 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-02-17 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5LQD
 
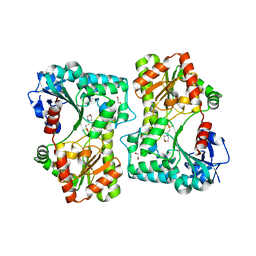 | | Trehalose-6-phosphate synthase, GDP-glucose-dependent OtsA | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha,alpha-trehalose-phosphate synthase | | Authors: | Miah, F, Asencion Diez, M.D, Stevenson, C.E.M, Lawson, D.M, Iglesias, A.A, Bornemann, S. | | Deposit date: | 2016-08-16 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Production and Utilization of GDP-glucose in the Biosynthesis of Trehalose 6-Phosphate by Streptomyces venezuelae.
J. Biol. Chem., 292, 2017
|
|
5LGV
 
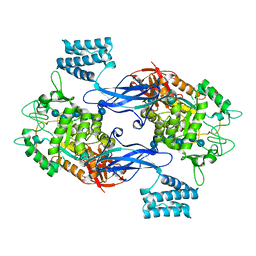 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant soaked in maltooctaose | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Mia, F, Barclay, J.E, Tang, M, Gorelik, A, Rashid, A.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2016-07-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-bound structures and site-directed mutagenesis identify the acceptor and secondary binding sites of Streptomyces coelicolor maltosyltransferase GlgE.
J.Biol.Chem., 291, 2016
|
|
