5BNJ
 
 | | CDK8/CYCC IN COMPLEX WITH 8-{3-Chloro-5-[4-(1-methyl-1H-pyrazol-4-yl)-phenyl]-pyridin- 4-yl}-2,8-diaza-spiro[4.5]decan-1-one | | Descriptor: | 8-{3-chloro-5-[4-(1-methyl-1H-pyrazol-4-yl)phenyl]pyridin-4-yl}-2,8-diazaspiro[4.5]decan-1-one, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Musil, D, Blagg, J, Wienke, D. | | Deposit date: | 2015-05-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | A selective chemical probe for exploring the role of CDK8 and CDK19 in human disease.
Nat.Chem.Biol., 11, 2015
|
|
8OKB
 
 | | SARS-CoV2 NSP5 in complex with a peptidomimetic ligand | | Descriptor: | 3C-like proteinase nsp5, methyl (4~{S})-4-[[(2~{S})-4-methyl-2-(phenylmethoxycarbonylamino)pentanoyl]amino]-5-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Calderone, V. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Development of a GC-376 Based Peptidomimetic PROTAC as a Degrader of 3-Chymotrypsin-like Protease of SARS-CoV-2.
Acs Med.Chem.Lett., 15, 2024
|
|
8OKC
 
 | | SARS-CoV2 NSP5 in complex with a GC-376 based peptidomimetic PROTAC | | Descriptor: | (phenylmethyl) ~{N}-[(2~{R})-1-[[(~{Z},2~{S})-5-[4-[[1-[2-[(3~{R})-2,6-bis(oxidanylidene)piperidin-3-yl]-6-fluoranyl-1,3-bis(oxidanylidene)isoindol-5-yl]piperidin-4-yl]methyl]piperazin-1-yl]-5-oxidanylidene-1-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]pent-3-en-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase nsp5 | | Authors: | Calderone, V. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of a GC-376 Based Peptidomimetic PROTAC as a Degrader of 3-Chymotrypsin-like Protease of SARS-CoV-2.
Acs Med.Chem.Lett., 15, 2024
|
|
4ID1
 
 | |
4GVM
 
 | |
6NVX
 
 | | Crystal structure of penicillin G acylase from Bacillus sp. FJAT-27231 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Blankenfeldt, W. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structures and protein engineering of three different penicillin G acylases from Gram-positive bacteria with different thermostability.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
4GW6
 
 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor | | Descriptor: | (2S)-[6-bromo-4-(4-chlorophenyl)-2-methylquinolin-3-yl](tert-butoxy)ethanoic acid, ARSENIC, Gag-Pol polyprotein | | Authors: | Feng, L, Kvaratskhelia, M. | | Deposit date: | 2012-08-31 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The A128T Resistance Mutation Reveals Aberrant Protein Multimerization as the Primary Mechanism of Action of Allosteric HIV-1 Integrase Inhibitors.
J.Biol.Chem., 288, 2013
|
|
6NVW
 
 | | Crystal structure of penicillin G acylase from Bacillus megaterium | | Descriptor: | CALCIUM ION, Penicillin G acylase | | Authors: | Blankenfeldt, W. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal structures and protein engineering of three different penicillin G acylases from Gram-positive bacteria with different thermostability.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
2ROY
 
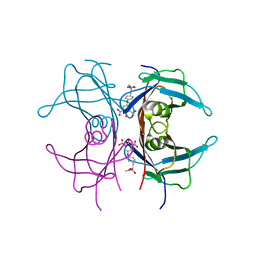 | | TRANSTHYRETIN (ALSO CALLED PREALBUMIN) COMPLEX WITH 3',5'-DINITRO-N-ACETYL-L-THYRONINE | | Descriptor: | 3',5'-DINITRO-N-ACETYL-L-THYRONINE, TRANSTHYRETIN | | Authors: | Wojtczak, A, Cody, V, Luft, J.R, Pangborn, W. | | Deposit date: | 1996-10-23 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of human transthyretin complexed with thyroxine at 2.0 A resolution and 3',5'-dinitro-N-acetyl-L-thyronine at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2ROX
 
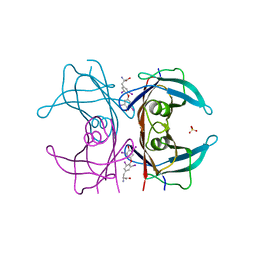 | | TRANSTHYRETIN (ALSO CALLED PREALBUMIN) COMPLEX WITH THYROXINE (T4) | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, SULFATE ION, TRANSTHYRETIN | | Authors: | Wojtczak, A, Cody, V, Luft, J.R, Pangborn, W. | | Deposit date: | 1996-10-23 | | Release date: | 1997-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of human transthyretin complexed with thyroxine at 2.0 A resolution and 3',5'-dinitro-N-acetyl-L-thyronine at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1K5D
 
 | | Crystal structure of Ran-GPPNHP-RanBP1-RanGAP complex | | Descriptor: | GTP-binding nuclear protein RAN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Seewald, M.J, Koerner, C, Wittinghofer, A, Vetter, I.R. | | Deposit date: | 2001-10-10 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RanGAP mediates GTP hydrolysis without an arginine finger.
Nature, 415, 2002
|
|
1K5G
 
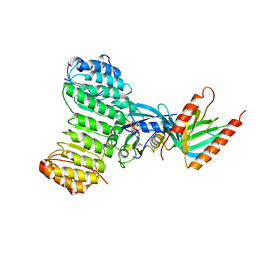 | | Crystal structure of Ran-GDP-AlFx-RanBP1-RanGAP complex | | Descriptor: | ALUMINUM FLUORIDE, GTP-binding nuclear protein RAN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Seewald, M.J, Koerner, C, Wittinghofer, A, Vetter, I.R. | | Deposit date: | 2001-10-10 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | RanGAP mediates GTP hydrolysis without an arginine finger.
Nature, 415, 2002
|
|
1RVD
 
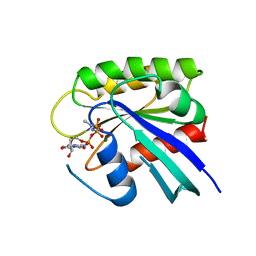 | | H-RAS COMPLEXED WITH DIAMINOBENZOPHENONE-BETA,GAMMA-IMIDO-GTP | | Descriptor: | 3-AMINOBENZOPHENONE-4-YL-AMINOHYDROXYPHOSPHINYLAMINOPHOSPHONIC ACID-GUANYLATE ESTER, MAGNESIUM ION, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Ahmadian, M.R, Zor, T, Vogt, D, Kabsch, W, Selinger, Z, Wittinghofer, A, Scheffzek, K. | | Deposit date: | 1999-05-03 | | Release date: | 1999-05-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Guanosine triphosphatase stimulation of oncogenic Ras mutants.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7D69
 
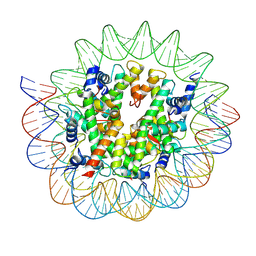 | | Cryo-EM structure of the nucleosome containing Giardia histones | | Descriptor: | 601L DNA (145-MER), Histone H2A, Histone H2B, ... | | Authors: | Sato, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Cryo-EM structure of the nucleosome core particle containing Giardia lamblia histones.
Nucleic Acids Res., 49, 2021
|
|
4JHQ
 
 | | Crystal structure of avidin - 6-(6-biotinamidohexanamido)hexanoylferrocene complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin, [(1,2,3,4,5-eta)-cyclopentadienyl][(1,2,3,4,5-eta)-{6-[(6-{[5-(2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanoyl]amino}hexanoyl)amino]hexanoyl}cyclopentadienyl]iron | | Authors: | Strzelczyk, P, Bujacz, A, Bujacz, G. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Ferrocene-Biotin Conjugates Targeting Cancer Cells: Synthesis, Interaction with Avidin, Cytotoxic Properties and the Crystal Structure of the Complex of Avidin with a Biotin-Linker-Ferrocene Conjugate
Organometallics, 32, 2013
|
|
4O0J
 
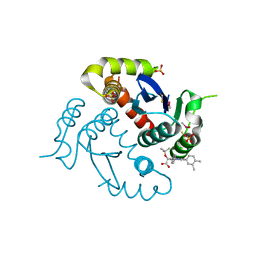 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor (2S)-tert-butoxy[4-(4-chlorophenyl)-6-(3,4-dimethylphenyl)-2,5-dimethylpyridin-3-yl]ethanoic acid | | Descriptor: | (2S)-tert-butoxy[4-(4-chlorophenyl)-6-(3,4-dimethylphenyl)-2,5-dimethylpyridin-3-yl]ethanoic acid, Integrase, SULFATE ION | | Authors: | Feng, L, Kvaratskhelia, M. | | Deposit date: | 2013-12-13 | | Release date: | 2014-07-02 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A New Class of Multimerization Selective Inhibitors of HIV-1 Integrase.
Plos Pathog., 10, 2014
|
|
6NVY
 
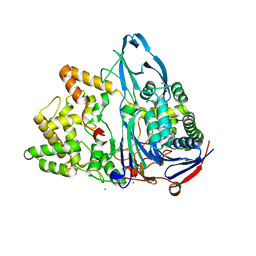 | | Crystal structure of penicillin G acylase from Bacillus thermotolerans | | Descriptor: | CALCIUM ION, GLYCEROL, Penicillin G acylase | | Authors: | Blankenfeldt, W. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structures and protein engineering of three different penicillin G acylases from Gram-positive bacteria with different thermostability.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
8PWZ
 
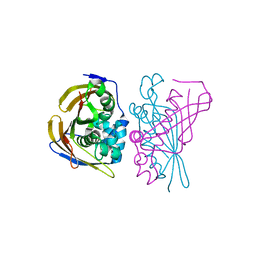 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadBD from Mycobacterium tuberculosis | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadB, UPF0336 protein Rv0504c | | Authors: | Rima, J, Grimoire, Y, Bories, P, Bardou, F, Quemard, A, Bon, C, Mourey, L, Tranier, S. | | Deposit date: | 2023-07-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.00196719 Å) | | Cite: | HadBD dehydratase from Mycobacterium tuberculosis fatty acid synthase type II: A singular structure for a unique function.
Protein Sci., 33, 2024
|
|
2A0K
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.8 A resolution | | Descriptor: | GLYCEROL, Nucleoside 2-deoxyribosyltransferase, SULFATE ION | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-06-16 | | Release date: | 2005-07-26 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
1TLM
 
 | |
2F67
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.6 A resolution with BENZO[CD]INDOL-2(1H)-ONE bound | | Descriptor: | BENZO[CD]INDOL-2(1H)-ONE, GLYCEROL, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-28 | | Release date: | 2005-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
2F62
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.5 A resolution with (2-ETHYLPHENYL)METHANOL bound | | Descriptor: | (2-ETHYLPHENYL)METHANOL, GLYCEROL, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-28 | | Release date: | 2005-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
2F2T
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.7 A resolution with 5-Aminoisoquinoline bound | | Descriptor: | GLYCEROL, ISOQUINOLIN-5-AMINE, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-17 | | Release date: | 2005-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
2F64
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.6 A resolution with 1-METHYLQUINOLIN-2(1H)-ONE bound | | Descriptor: | 1-METHYLQUINOLIN-2(1H)-ONE, GLYCEROL, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-28 | | Release date: | 2005-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
3BJE
 
 | |
