4IP3
 
 | | Complex structure of OspI and Ubc13 | | Descriptor: | ORF169b, Ubiquitin-conjugating enzyme E2 N | | Authors: | Fu, P, Jin, M, Zhang, X, Xu, L, Xia, Z, Zhu, Y. | | Deposit date: | 2013-01-09 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure Analysis of Ubc13 Inactivation
To be Published
|
|
5H1L
 
 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with 7-nt U4 snRNA fragment | | Descriptor: | GLYCEROL, Gem-associated protein 5, U4 snRNA (5'-R(*AP*UP*UP*UP*UP*UP*G)-3') | | Authors: | Jin, W, Wang, Y, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
5GW4
 
 | | Structure of Yeast NPP-TRiC | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Zang, Y, Jin, M, Wang, H, Cong, Y. | | Deposit date: | 2016-09-08 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5GW5
 
 | | Structure of TRiC-AMP-PNP | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Zang, Y, Jin, M, Wang, H, Cong, Y. | | Deposit date: | 2016-09-08 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
8X6M
 
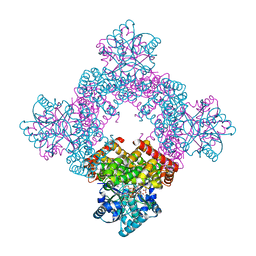 | | Crystal Structure of Glycerol Dehydrogenase in the Presence of NAD+ and Glycerol | | Descriptor: | GLYCEROL, Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, T, Kang, J.Y, Jin, M, Yang, J, Kim, H, Noh, C, Eom, S.H. | | Deposit date: | 2023-11-21 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the octamerization of glycerol dehydrogenase.
Plos One, 19, 2024
|
|
7CLT
 
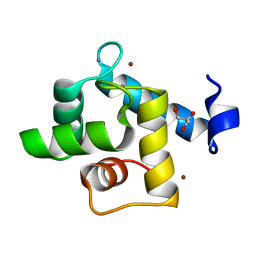 | | Crystal structure of the EFhd1/Swiprosin-2, a mitochondrial actin-binding protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D1, GLYCEROL, ... | | Authors: | Mun, S.A, Park, J, Park, K.R, Lee, Y, Kang, J.Y, Park, T, Jin, M, Yang, J, Jun, C.D, Eom, S.H. | | Deposit date: | 2020-07-22 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07380986 Å) | | Cite: | Structural and Biochemical Characterization of EFhd1/Swiprosin-2, an Actin-Binding Protein in Mitochondria.
Front Cell Dev Biol, 8, 2020
|
|
7YGW
 
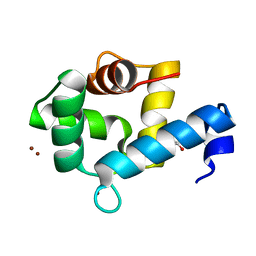 | | Crystal structure of the Zn2+-bound EFhd1/Swiprosin-2 | | Descriptor: | EF-hand domain-containing protein D1, GLYCEROL, ZINC ION | | Authors: | Mun, S.A, Park, J, Kang, J.Y, Park, T, Jin, M, Yang, J, Eom, S.H. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural and biochemical insights into Zn 2+ -bound EF-hand proteins, EFhd1 and EFhd2.
Iucrj, 10, 2023
|
|
7YGV
 
 | | Crystal structure of the Ca2+-bound EFhd1/Swiprosin-2 | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D1, GLYCEROL, ... | | Authors: | Mun, S.A, Park, J, Kang, J.Y, Park, T, Jin, M, Ynag, J, Eom, S.H. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical insights into Zn 2+ -bound EF-hand proteins, EFhd1 and EFhd2.
Iucrj, 10, 2023
|
|
7YGY
 
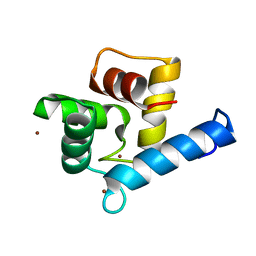 | | Crystal structure of the Zn2+-bound EFhd2/Swiprosin-1 | | Descriptor: | EF-hand domain-containing protein D2, ZINC ION | | Authors: | Mun, S.A, Park, J, Kang, J.Y, Park, T, Jin, M, Yang, J, Eom, S.H. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical insights into Zn 2+ -bound EF-hand proteins, EFhd1 and EFhd2.
Iucrj, 10, 2023
|
|
6QZK
 
 | | Structure of Clostridium butyricum Argonaute bound to a guide DNA (5' deoxycytidine) and a 19-mer target DNA | | Descriptor: | Clostridium butyricum Argonaute, DNA target (5'-D(T*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*T)-3'), FORMIC ACID, ... | | Authors: | Swarts, D.C, Jinek, M, Hegge, J.W, Van der Oost, J. | | Deposit date: | 2019-03-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.548 Å) | | Cite: | DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute.
Nucleic Acids Res., 47, 2019
|
|
6F9W
 
 | |
6F9N
 
 | | CRYSTAL STRUCTURE OF THE HUMAN CPSF160-WDR33 COMPLEX | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, pre-mRNA 3' end processing protein WDR33 | | Authors: | Clerici, M, Jinek, M. | | Deposit date: | 2017-12-14 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the assembly and polyA signal recognition mechanism of the human CPSF complex.
Elife, 6, 2017
|
|
6FUW
 
 | | Cryo-EM structure of the human CPSF160-WDR33-CPSF30 complex bound to the PAS AAUAAA motif at 3.1 Angstrom resolution | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, RNA (5'-R(P*AP*AP*UP*AP*AP*AP*GP*G)-3'), ... | | Authors: | Clerici, M, Faini, M, Jinek, M. | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of AAUAAA polyadenylation signal recognition by the human CPSF complex.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8Q40
 
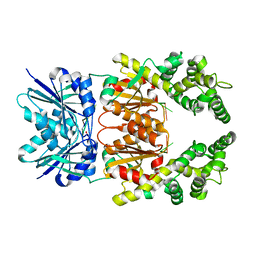 | | Crystal structure of cA4 activated Can2 in complex with a cleaved DNA substrate | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*TP*CP*A)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q42
 
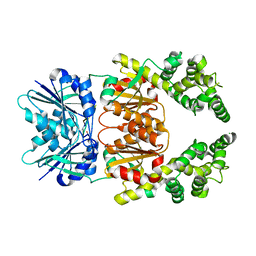 | | Crystal structure of cA4-bound Can2 (E341A) in complex with oligo-A DNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*AP*AP*AP*A)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q41
 
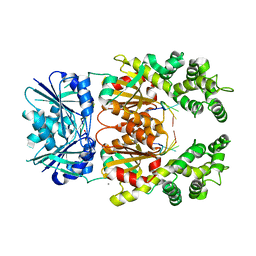 | | Crystal structure of Can2 (E341A) bound to cA4 and TTTAAA ssDNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*TP*TP*AP*AP*A)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q43
 
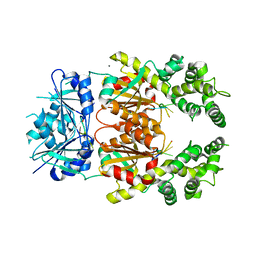 | | Crystal structure of cA4-bound Can2 (E341A) in complex with oligo-C DNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*CP*CP*CP*CP*C)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q44
 
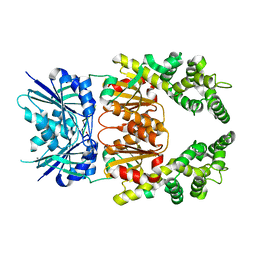 | | Crystal structure of cA4-bound Can2 (E364R) in complex with oligo-T DNA | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), DNA (5'-D(*TP*TP*T)-3'), DUF1887 family protein, ... | | Authors: | Jungfer, K, Sigg, A, Jinek, M. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Nucleic Acids Res., 52, 2024
|
|
8Q3Z
 
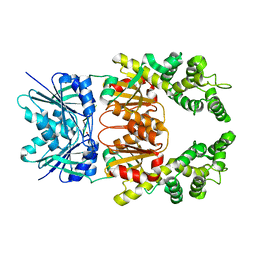 | |
8Q3Y
 
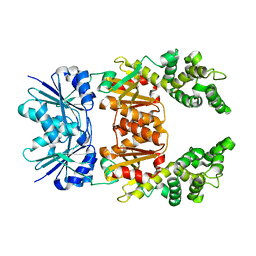 | |
8QLP
 
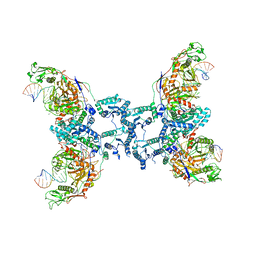 | | CryoEM structure of the RNA/DNA bound SPARTA (BabAgo/TIR-APAZ) tetrameric complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*AP*TP*TP*AP*GP*AP*GP*CP*CP*GP*TP*C)-3'), MAGNESIUM ION, RNA (5'-R(*AP*UP*GP*AP*CP*GP*GP*CP*UP*CP*UP*AP*AP*UP*CP*UP*AP*UP*UP*AP*GP*U)-3'), ... | | Authors: | Finocchio, G, Koopal, B, Potocnik, A, Heijstek, C, Jinek, M, Swarts, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Target DNA-dependent activation mechanism of the prokaryotic immune system SPARTA.
Nucleic Acids Res., 52, 2024
|
|
8QLO
 
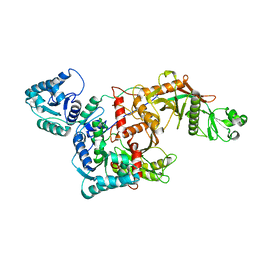 | | CryoEM structure of the apo SPARTA (BabAgo/TIR-APAZ) complex | | Descriptor: | Short prokaryotic Argonaute, Toll/interleukin-1 receptor domain-containing protein | | Authors: | Finocchio, G, Koopal, B, Potocnik, A, Heijstek, C, Jinek, M, Swarts, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Target DNA-dependent activation mechanism of the prokaryotic immune system SPARTA.
Nucleic Acids Res., 52, 2024
|
|
3SGB
 
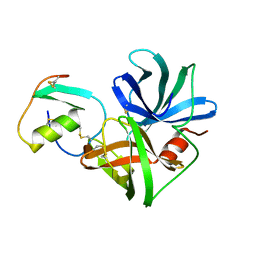 | | STRUCTURE OF THE COMPLEX OF STREPTOMYCES GRISEUS PROTEASE B AND THE THIRD DOMAIN OF THE TURKEY OVOMUCOID INHIBITOR AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | PROTEINASE B (SGPB), TURKEY OVOMUCOID INHIBITOR (OMTKY3) | | Authors: | Read, R.J, Fujinaga, M, Sielecki, A.R, James, M.N.G. | | Deposit date: | 1983-01-21 | | Release date: | 1983-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the complex of Streptomyces griseus protease B and the third domain of the turkey ovomucoid inhibitor at 1.8-A resolution.
Biochemistry, 22, 1983
|
|
7OX8
 
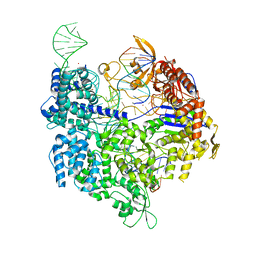 | | Target-bound SpCas9 complex with TRAC full RNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OXA
 
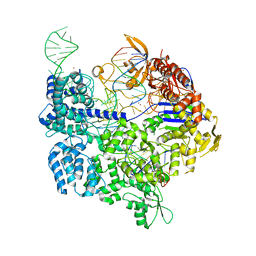 | | Target-bound SpCas9 complex with AAVS1 chimeric RNA-DNA guide | | Descriptor: | AAVS1 non-target DNA strand, AAVS1 target DNA strand, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
