4IN0
 
 | |
4I5L
 
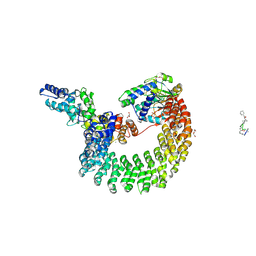 | | Structural mechanism of trimeric PP2A holoenzyme involving PR70: insight for Cdc6 dephosphorylation | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, MALONATE ION, ... | | Authors: | Wlodarchak, N, Satyshur, K.A, Guo, F, Xing, Y. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of the Ca(2+)-dependent PP2A heterotrimer and insights into Cdc6 dephosphorylation.
Cell Res., 23, 2013
|
|
1XNS
 
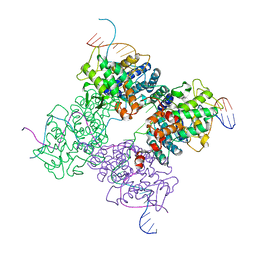 | | Peptide trapped Holliday junction intermediate in Cre-loxP recombination | | Descriptor: | Recombinase CRE, loxP DNA | | Authors: | Ghosh, K, Lau, C.K, Guo, F, Segall, A.M, Van Duyne, G.D. | | Deposit date: | 2004-10-05 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Peptide trapping of the Holliday junction intermediate in Cre-loxP site-specific recombination.
J.Biol.Chem., 280, 2005
|
|
1XO0
 
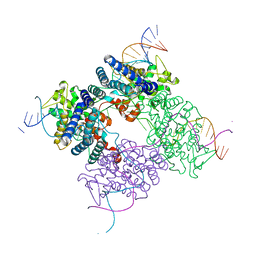 | | High resolution structure of the holliday junction intermediate in cre-loxp site-specific recombination | | Descriptor: | Recombinase CRE, loxP | | Authors: | Ghosh, K, Lau, C.K, Guo, F, Segall, A.M, Van Duyne, G.D. | | Deposit date: | 2004-10-05 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Peptide trapping of the Holliday junction intermediate in Cre-loxP site-specific recombination.
J.Biol.Chem., 280, 2005
|
|
3GIX
 
 | |
3JCI
 
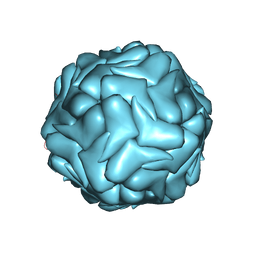 | | 2.9 Angstrom Resolution Cryo-EM 3-D Reconstruction of Close-packed PCV2 Virus-like Particles | | Descriptor: | Capsid protein | | Authors: | Liu, Z, Guo, F, Wang, F, Li, T.C, Jiang, W. | | Deposit date: | 2015-12-13 | | Release date: | 2016-02-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | 2.9 angstrom Resolution Cryo-EM 3D Reconstruction of Close-Packed Virus Particles.
Structure, 24, 2016
|
|
7TGH
 
 | | Cryo-EM structure of respiratory super-complex CI+III2 from Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Zhou, L, Maldonado, M, Padavannil, A, Guo, F, Letts, J.A. | | Deposit date: | 2022-01-07 | | Release date: | 2022-04-06 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of Tetrahymena 's respiratory chain reveal the diversity of eukaryotic core metabolism.
Science, 376, 2022
|
|
6B4S
 
 | |
6N5P
 
 | | Structure of Human pir-miRNA-340 Apical Loop and One-base-pair Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
6N5K
 
 | | Structure of Human pir-miRNA-449c Apical Loop and One-base-pair Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
6N5O
 
 | | Structure of Human pir-miRNA-202 Apical Loop and One-base-pair Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
6N5Q
 
 | | Structure of Human pir-miRNA-378a Apical Loop and One-base-pair Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.946 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
6N5S
 
 | | Structure of Human pir-miRNA-320b-2 Apical Loop and One-base-pair Stem Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
6N5N
 
 | | Structure of Human pir-miRNA-208a Apical Loop and One-base-pair Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
6N5T
 
 | | Structure of Human pir-miRNA-378a Apical Loop Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.787 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
8HDK
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (minor class in symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-11-04 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8HAY
 
 | | d4-bound btDPP4 | | Descriptor: | (1~{R})-1-[[4-[5-[[(1~{R})-6,7-dimethoxy-2-methyl-3,4-dihydro-1~{H}-isoquinolin-1-yl]methyl]-2-methoxy-phenoxy]phenyl]methyl]-6,7-dimethoxy-2-methyl-3,4-dihydro-1~{H}-isoquinoline, btDPP4 | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-10-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
4WOC
 
 | | Proteinase-K Post-Surface Acoustic Waves | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | French, J.B. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Precise Manipulation and Patterning of Protein Crystals for Macromolecular Crystallography Using Surface Acoustic Waves.
Small, 11, 2015
|
|
4WO9
 
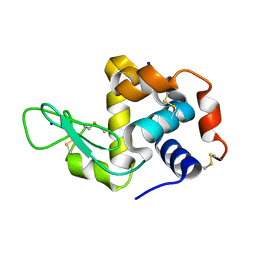 | | Lysozyme Post-Surface Acoustic Waves | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | French, J.B. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Precise Manipulation and Patterning of Protein Crystals for Macromolecular Crystallography Using Surface Acoustic Waves.
Small, 11, 2015
|
|
4WOB
 
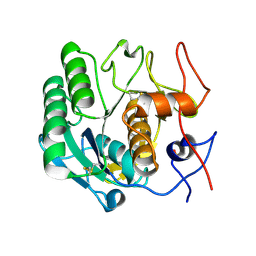 | | Proteinase-K Pre-Surface Acoustic Wave | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | French, J.B. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Precise Manipulation and Patterning of Protein Crystals for Macromolecular Crystallography Using Surface Acoustic Waves.
Small, 11, 2015
|
|
4WO6
 
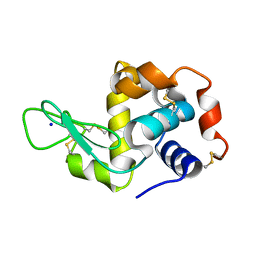 | | Lysozyme Pre-surface acoustic wave | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | French, J.B. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Precise Manipulation and Patterning of Protein Crystals for Macromolecular Crystallography Using Surface Acoustic Waves.
Small, 11, 2015
|
|
4WOA
 
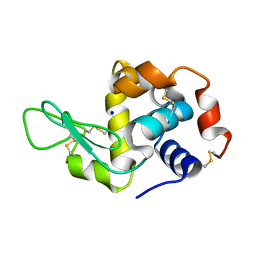 | |
4FHZ
 
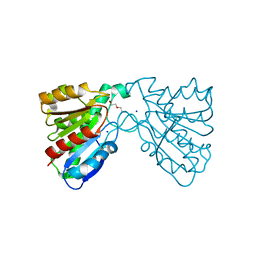 | | Crystal structure of a carboxyl esterase at 2.0 angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phospholipase/Carboxylesterase, SODIUM ION | | Authors: | Wu, L, Ma, J, Zhou, J, Yu, H. | | Deposit date: | 2012-06-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Enhanced enantioselectivity of a carboxyl esterase from Rhodobacter sphaeroides by directed evolution.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
5K8L
 
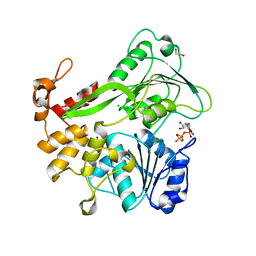 | | Crystal structure of ZIKV NS3 helicase in complex with GTP-gammar S | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Cao, X, Li, Y, Jin, T. | | Deposit date: | 2016-05-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Molecular mechanism of divalent-metal-induced activation of NS3 helicase and insights into Zika virus inhibitor design.
Nucleic Acids Res., 44, 2016
|
|
5K8U
 
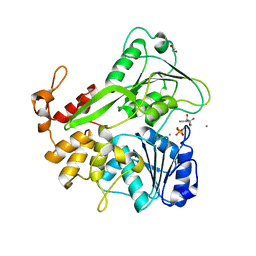 | | Crystal structure of ZIKV NS3 helicase in complex with ADP and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Cao, X, Li, Y, Jin, T. | | Deposit date: | 2016-05-31 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Molecular mechanism of divalent-metal-induced activation of NS3 helicase and insights into Zika virus inhibitor design.
Nucleic Acids Res., 44, 2016
|
|
