6LSR
 
 | | Cryo-EM structure of a pre-60S ribosomal subunit - state B | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | Deposit date: | 2020-01-20 | | Release date: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
6LSS
 
 | | Cryo-EM structure of a pre-60S ribosomal subunit - state preA | | Descriptor: | 28S rRNA, 5S rRNA, 60S ribosomal protein L11, ... | | Authors: | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | Deposit date: | 2020-01-20 | | Release date: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
5G06
 
 | | Cryo-EM structure of yeast cytoplasmic exosome | | Descriptor: | EXOSOME COMPLEX COMPONENT CSL4, EXOSOME COMPLEX COMPONENT MTR3, EXOSOME COMPLEX COMPONENT RRP4, ... | | Authors: | Liu, J.J, Niu, C.Y, Wu, Y, Tan, D, Wang, Y, Ye, M.D, Liu, Y, Zhao, W.W, Zhou, K, Liu, Q.S, Dai, J.B, Yang, X.R, Dong, M.Q, Huang, N, Wang, H.W. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryoem Structure of Yeast Cytoplasmic Exosome Complex.
Cell Res., 26, 2016
|
|
8BAL
 
 | |
8BBL
 
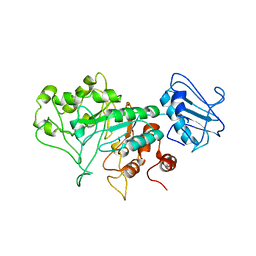 | | SGL a GH20 family sulfoglycosidase | | Descriptor: | Beta-N-acetylhexosaminidase | | Authors: | Dong, M.D, Roth, C.R, Jin, Y.J. | | Deposit date: | 2022-10-13 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 2022
|
|
7S0Y
 
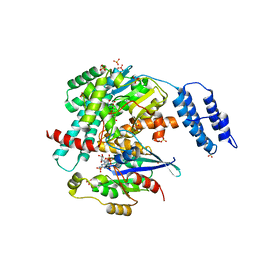 | | Structures of TcdB in complex with Cdc42 | | Descriptor: | Cell division control protein 42 homolog, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zheng, L, Rongsheng, J, Peng, C. | | Deposit date: | 2021-08-31 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis for selective modification of Rho and Ras GTPases by Clostridioides difficile toxin B.
Sci Adv, 7, 2021
|
|
7S0Z
 
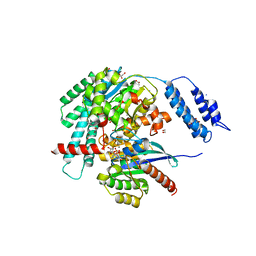 | | Structures of TcdB in complex with R-Ras | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, AMMONIUM ION, ... | | Authors: | Zheng, L, Rongsheng, J, Peng, C. | | Deposit date: | 2021-08-31 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for selective modification of Rho and Ras GTPases by Clostridioides difficile toxin B.
Sci Adv, 7, 2021
|
|
3ZEE
 
 | | Electron cyro-microscopy helical reconstruction of Par-3 N terminal domain | | Descriptor: | PARTITIONING DEFECTIVE 3 HOMOLOG | | Authors: | Zhang, Y, Wang, W, Chen, J, Zhang, K, Gao, F, Gong, W, Zhang, M, Sun, F, Feng, W. | | Deposit date: | 2012-12-05 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural Insights Into the Intrinsic Self-Assembly of Par-3 N-Terminal Domain.
Structure, 21, 2013
|
|
2VXR
 
 | |
8C8G
 
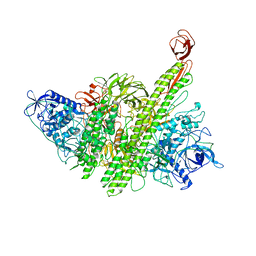 | |
8CQ2
 
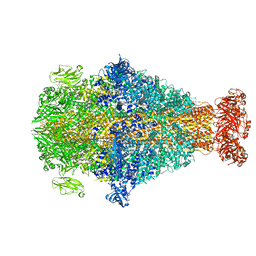 | | Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, C16S, C20S, C870S, T1279C mutant | | Descriptor: | TcdA1 | | Authors: | Nganga, P.N, Roderer, D, Belyy, A, Prumbaum, D, Raunser, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Kinetics of the syringe-like injection mechanism of Tc toxins
to be published
|
|
8CPZ
 
 | | Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, K1179W mutant | | Descriptor: | TcdA1 | | Authors: | Nganga, P.N, Roderer, D, Belyy, A, Prumbaum, D, Raunser, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Kinetics of the syringe-like injection mechanism of Tc toxins
to be published
|
|
8CQ0
 
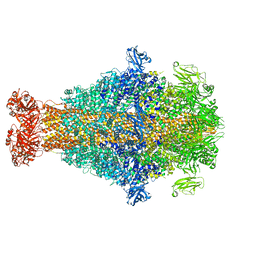 | | Photorhabdus luminescens TcdA1 prepore-to-pore intermediate, K567W K2008W mutant | | Descriptor: | TcdA1 | | Authors: | Nganga, P.N, Roderer, D, Belyy, A, Prumbaum, D, Raunser, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Kinetics of the syringe-like injection mechanism of Tc toxins
to be published
|
|
6RIM
 
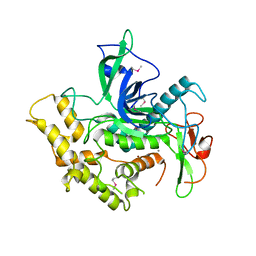 | |
5Z2C
 
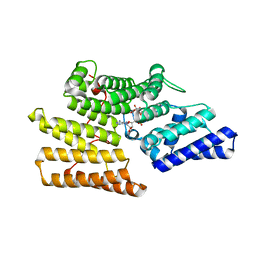 | | Crystal structure of ALPK-1 N-terminal domain in complex with ADP-heptose | | Descriptor: | Alpha-protein kinase 1, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4S,5S,6R)-6-[(1S)-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Ding, J, She, Y, Shao, F. | | Deposit date: | 2018-01-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Alpha-kinase 1 is a cytosolic innate immune receptor for bacterial ADP-heptose.
Nature, 561, 2018
|
|
5L21
 
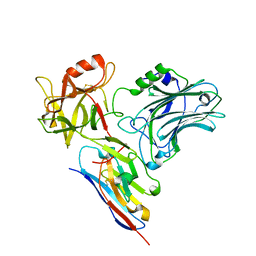 | |
5LR0
 
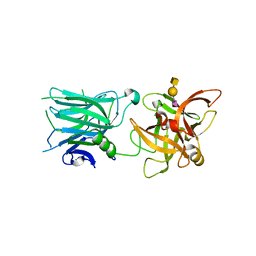 | | Binding domain of Botulinum Neurotoxin DC in complex with SialylT | | Descriptor: | Botulinum neurotoxin D/C protein, N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Berntsson, R.P.-A, Stenmark, P. | | Deposit date: | 2016-08-18 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis for the unique ganglioside and cell membrane recognition mechanism of botulinum neurotoxin DC.
Nat Commun, 8, 2017
|
|
9BKJ
 
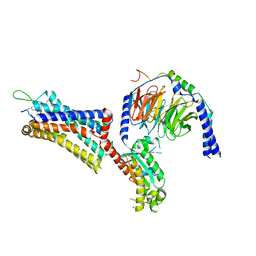 | | Cholecystokinin 1 receptor (CCK1R) Y140A mutant, Gq chimera (mGsqi) complex | | Descriptor: | AMINO GROUP, Cholecystokinin receptor type A, Cholecystokinin-8, ... | | Authors: | Cary, B.P, Harikumar, K.G, Zhao, P, Desai, A.J, Mobbs, J.M, Toufaily, C, Furness, S.G.B, Christopoulos, A, Belousoff, M.J, Wootten, D, Sexton, P.M, Miller, L.J. | | Deposit date: | 2024-04-29 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Cholesterol-dependent dynamic changes in the conformation of the type 1 cholecystokinin receptor affect ligand binding and G protein coupling.
Plos Biol., 22, 2024
|
|
9BKK
 
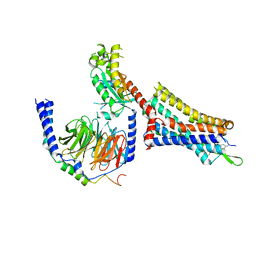 | | Cholecystokinin 1 receptor (CCK1R) sterol 7M mutant, Gq chimera (mGsqi) complex | | Descriptor: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Harikumar, K.G, Zhao, P, Cary, B.P, Xu, X, Desai, A.J, Mobbs, J.I, Toufaily, C, Furness, S.G.B, Christopoulos, A, Belousoff, M.J, Wootten, D, Sexton, P.M, Miller, L.J. | | Deposit date: | 2024-04-29 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Cholesterol-dependent dynamic changes in the conformation of the type 1 cholecystokinin receptor affect ligand binding and G protein coupling.
Plos Biol., 22, 2024
|
|
3U28
 
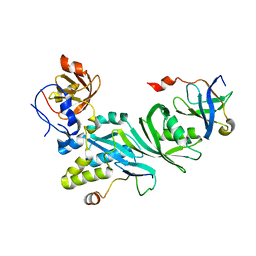 | | Crystal structure of a Cbf5-Nop10-Gar1 complex from Saccharomyces cerevisiae | | Descriptor: | H/ACA ribonucleoprotein complex subunit 1, H/ACA ribonucleoprotein complex subunit 3, H/ACA ribonucleoprotein complex subunit 4 | | Authors: | Ye, K, Li, S. | | Deposit date: | 2011-10-02 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase
Genes Dev., 25, 2011
|
|
7U1Z
 
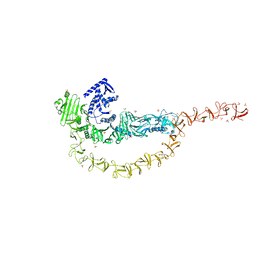 | | Crystal structure of the DRBD and CROPs of TcdA | | Descriptor: | SULFATE ION, Toxin A | | Authors: | Baohua, C, Peng, C, Kay, P, Rongsheng, J. | | Deposit date: | 2022-02-22 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure and conformational dynamics of Clostridioides difficile toxin A.
Life Sci Alliance, 5, 2022
|
|
7UIB
 
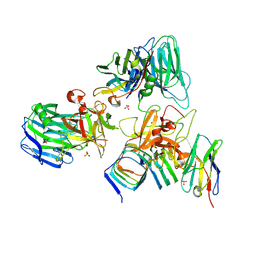 | | Crystal structure of BoNT/E receptor binding domain in complex with SV2, VHH, and sialic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-beta-neuraminic acid, ... | | Authors: | Liu, Z, Jin, R, Chen, P. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2.
Nat Commun, 14, 2023
|
|
7UIA
 
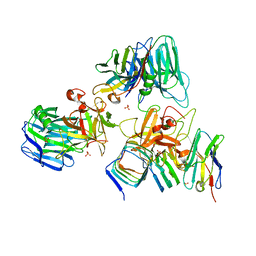 | | Crystal structure of BoNT/E receptor binding domain in complex with SV2 and VHH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, Z, Jin, R, Chen, P. | | Deposit date: | 2022-03-28 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2.
Nat Commun, 14, 2023
|
|
7UIE
 
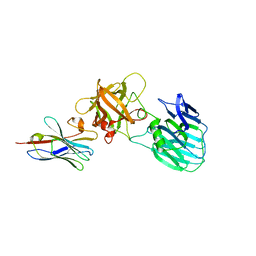 | | Crystal structure of HcE-JLE-G6 | | Descriptor: | Botulinum neurotoxin E heavy chain, JLE-G6 | | Authors: | Jin, R, Lam, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2.
Nat Commun, 14, 2023
|
|
4I6P
 
 | | Crystal structure of Par3-NTD domain | | Descriptor: | Partitioning defective 3 homolog | | Authors: | Wang, W, Gao, F, Gong, W, Sun, F, Feng, W. | | Deposit date: | 2012-11-29 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the intrinsic self-assembly of par-3 N-terminal domain.
Structure, 21, 2013
|
|
