1O0X
 
 | |
1O51
 
 | |
1O59
 
 | |
6W2D
 
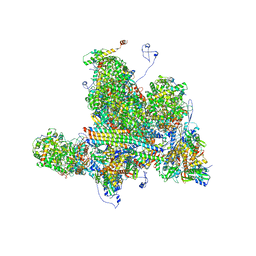 | | Structures of Capsid and Capsid-Associated Tegument Complex inside the Epstein-Barr Virus | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Liu, W, Cui, Y.X, Wang, C.Y, Li, Z.H, Gong, D.Y, Dai, X.H, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2020-03-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of capsid and capsid-associated tegument complex inside the Epstein-Barr virus.
Nat Microbiol, 5, 2020
|
|
6W19
 
 | | Structures of Capsid and Capsid-Associated Tegument Complex inside the Epstein-Barr Virus | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Liu, W, Cui, Y.X, Wang, C.Y, Li, Z.H, Gong, D.Y, Dai, X.H, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2020-03-03 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structures of capsid and capsid-associated tegument complex inside the Epstein-Barr virus.
Nat Microbiol, 5, 2020
|
|
6W2E
 
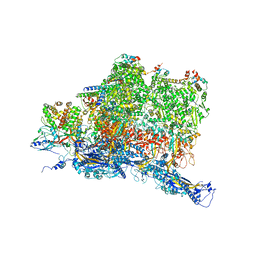 | | Structures of Capsid and Capsid-Associated Tegument Complex inside the Epstein-Barr Virus | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Liu, W, Cui, Y.X, Wang, C.Y, Li, Z.H, Gong, D.Y, Dai, X.H, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2020-03-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures of capsid and capsid-associated tegument complex inside the Epstein-Barr virus.
Nat Microbiol, 5, 2020
|
|
1O1X
 
 | |
1O1Y
 
 | |
1O5H
 
 | |
1O3U
 
 | |
1O22
 
 | |
1O4V
 
 | |
1O1Z
 
 | |
1O2D
 
 | |
1O4U
 
 | |
8D35
 
 | |
6ODM
 
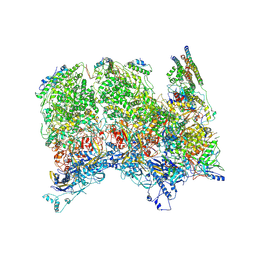 | | Herpes simplex virus type 1 (HSV-1) portal vertex-adjacent capsid/CATC, asymmetric unit | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Liu, Y.T, Jih, J, Dai, X.H, Bi, G.Q, Zhou, Z.H. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures of herpes simplex virus type 1 portal vertex and packaged genome.
Nature, 570, 2019
|
|
1VLR
 
 | |
5EJM
 
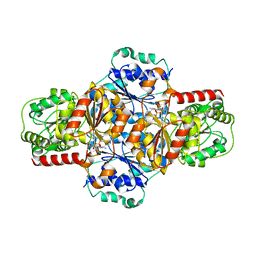 | | ThDP-Mn2+ complex of R413A variant of EcMenD soaked with 2-ketoglutarate for 35 min | | Descriptor: | (4~{R})-4-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-3-ium-2-yl]-4-oxidanyl-butanoic acid, 1,2-ETHANEDIOL, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ... | | Authors: | Song, H.G, Dong, C, Chen, Y.Z, Sun, Y.R, Guo, Z.H. | | Deposit date: | 2015-11-02 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Two active site arginines are critical determinants of substrate binding and catalysis in MenD, a thiamine-dependent enzyme in menaquinone biosynthesis.
Biochem. J., 2018
|
|
5LBV
 
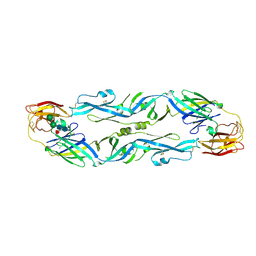 | | Structural basis of zika and dengue virus potent antibody cross-neutralization | | Descriptor: | SODIUM ION, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, envelope protein E | | Authors: | Barba-Spaeth, G. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of potent Zika-dengue virus antibody cross-neutralization.
Nature, 536, 2016
|
|
5LBS
 
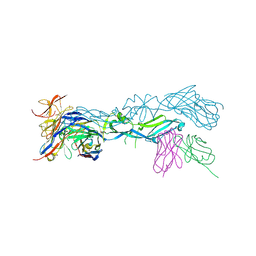 | | structural basis of Zika and Dengue virus potent antibody cross-neutralization | | Descriptor: | 1,2-ETHANEDIOL, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8, SULFATE ION, ... | | Authors: | Vaney, M.C, Rouvinski, A, Barba-Spaeth, G, Rey, F.A. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis of potent Zika-dengue virus antibody cross-neutralization.
Nature, 536, 2016
|
|
5LCV
 
 | |
5Z2R
 
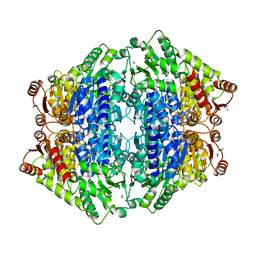 | | ThDP-Mn2+ complex of R395K variant of EcMenD soaked with 2-ketoglutarate for 5 min | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, FORMIC ACID, ... | | Authors: | Qin, M.M, Guo, Z.H. | | Deposit date: | 2018-01-03 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two active site arginines are critical determinants of substrate binding and catalysis in MenD: a thiamine-dependent enzyme in menaquinone biosynthesis.
Biochem. J., 475, 2018
|
|
5Z2P
 
 | | ThDP-Mn2+ complex of R413K variant of EcMenD soaked with 2-ketoglutarate for 5 min | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, FORMIC ACID, ... | | Authors: | Qin, M.M, Guo, Z.H. | | Deposit date: | 2018-01-03 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two active site arginines are critical determinants of substrate binding and catalysis in MenD: a thiamine-dependent enzyme in menaquinone biosynthesis.
Biochem. J., 475, 2018
|
|
5Z2U
 
 | | ThDP-Mn2+ complex of R395A variant of EcMenD soaked with 2-ketoglutarate for 5 min | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, 1,2-ETHANEDIOL, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ... | | Authors: | Qin, M.M, Guo, Z.H. | | Deposit date: | 2018-01-04 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Two active site arginines are critical determinants of substrate binding and catalysis in MenD: a thiamine-dependent enzyme in menaquinone biosynthesis.
Biochem. J., 475, 2018
|
|
