8I1J
 
 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP at pH 9.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1G
 
 | |
8I19
 
 | |
8I8T
 
 | |
8I1F
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 8.6 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I8S
 
 | |
2DQE
 
 | | Crystal structure of hyhel-10 FV mutant (Hy53a) complexed with hen egg lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Shiroishi, M, Kondo, H, Tsumoto, K, Kumagai, I. | | Deposit date: | 2006-05-25 | | Release date: | 2007-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural consequences of mutations in interfacial Tyr residues of a protein antigen-antibody complex. The case of HyHEL-10-HEL
J.Biol.Chem., 282, 2007
|
|
8IC7
 
 | | exo-beta-D-arabinofuranosidase ExoMA2 from Microbacterium arabinogalactanolyticum in complex with beta-D-arabinofuranose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Fukushima, R, Kashima, T, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8IC6
 
 | | exo-beta-D-arabinanase ExoMA2 from Microbacterium arabinogalactanolyticum in complex with Tris | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Fukushima, R, Kashima, T, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8IC8
 
 | | Exo-alpha-D-arabinofuranosidase from Microbacterium arabinogalactanolyticum | | Descriptor: | Exo-alpha-D-arabinofuranosidase, PHOSPHATE ION | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
2DQJ
 
 | | Crystal structure of hyhel-10 FV (wild-type) complexed with hen egg lysozyme at 1.8A resolution | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Shiroishi, M, Kondo, H, Tsumoto, K, Kumagai, I. | | Deposit date: | 2006-05-26 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural consequences of mutations in interfacial Tyr residues of a protein antigen-antibody complex. The case of HyHEL-10-HEL
J.Biol.Chem., 282, 2007
|
|
8WGT
 
 | | Crystal structure of V30M-TTR in complex with compound 7 | | Descriptor: | Transthyretin, [4,7-bis(chloranyl)-2-ethyl-1-benzofuran-3-yl]-[3,5-bis(iodanyl)-4-oxidanyl-phenyl]methanone | | Authors: | Yokoyama, T. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Development of Benziodarone Analogues with Enhanced Potency for Selective Binding to Transthyretin in Human Plasma.
J.Med.Chem., 67, 2024
|
|
8WGS
 
 | | Crystal structure of V30M-TTR in complex with compound 4 | | Descriptor: | Transthyretin, [3,5-bis(iodanyl)-4-oxidanyl-phenyl]-(2-ethyl-4-iodanyl-1-benzofuran-3-yl)methanone | | Authors: | Yokoyama, T. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Development of Benziodarone Analogues with Enhanced Potency for Selective Binding to Transthyretin in Human Plasma.
J.Med.Chem., 67, 2024
|
|
8WGU
 
 | | Crystal structure of V30M-TTR in complex with compound 20 | | Descriptor: | Transthyretin, [3,5-bis(iodanyl)-4-oxidanyl-phenyl]-[2-ethyl-4,7-bis(fluoranyl)-1-benzofuran-3-yl]methanone | | Authors: | Yokoyama, T. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.508 Å) | | Cite: | Development of Benziodarone Analogues with Enhanced Potency for Selective Binding to Transthyretin in Human Plasma.
J.Med.Chem., 67, 2024
|
|
7DFP
 
 | | Human dopamine D2 receptor in complex with spiperone | | Descriptor: | 8-[4-(4-fluorophenyl)-4-oxidanylidene-butyl]-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, D(2) dopamine receptor,Soluble cytochrome b562, FabH, ... | | Authors: | Im, D, Shimamura, T, Iwata, S. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the dopamine D 2 receptor in complex with the antipsychotic drug spiperone.
Nat Commun, 11, 2020
|
|
1GP5
 
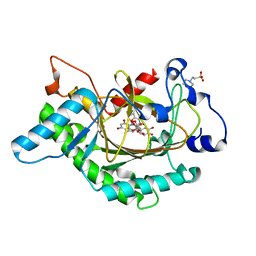 | | Anthocyanidin synthase from Arabidopsis thaliana complexed with trans-dihydroquercetin | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, (2S,3S)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Wilmouth, R.C, Turnbull, J.J, Welford, R.W.D, Clifton, I.J, Prescott, A.G, Schofield, C.J. | | Deposit date: | 2001-10-30 | | Release date: | 2002-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Mechanism of Anthocyanidin Synthase from Arabidopsis Thaliana.
Structure, 10, 2002
|
|
5C1T
 
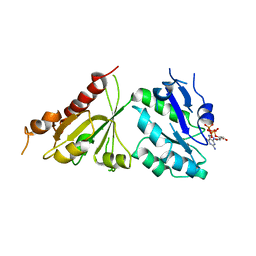 | | Crystal structure of the GTP-bound wild type EhRabX3 from Entamoeba histolytica | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Small GTPase EhRabX3 | | Authors: | Srivastava, V.K, Chandra, M, Datta, S. | | Deposit date: | 2015-06-15 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Crystal Structure Analysis of Wild Type and Fast Hydrolyzing Mutant of EhRabX3, a Tandem Ras Superfamily GTPase from Entamoeba histolytica.
J.Mol.Biol., 428, 2016
|
|
6LM0
 
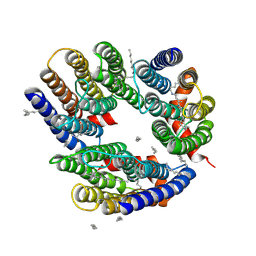 | | The crystal structure of cyanorhodopsin (CyR) N2098R from cyanobacteria Calothrix sp. NIES-2098 | | Descriptor: | DECANE, HEXANE, N-OCTANE, ... | | Authors: | Hosaka, T, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2019-12-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A unique clade of light-driven proton-pumping rhodopsins evolved in the cyanobacterial lineage.
Sci Rep, 10, 2020
|
|
5C1S
 
 | |
6LM1
 
 | | The crystal structure of cyanorhodopsin (CyR) N4075R from cyanobacteria Tolypothrix sp. NIES-4075 | | Descriptor: | DECANE, DODECANE, HEXADECANE, ... | | Authors: | Hosaka, T, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2019-12-24 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A unique clade of light-driven proton-pumping rhodopsins evolved in the cyanobacterial lineage.
Sci Rep, 10, 2020
|
|
1RMS
 
 | | CRYSTAL STRUCTURES OF RIBONUCLEASE MS COMPLEXED WITH 3'-GUANYLIC ACID A GP*C ANALOGUE, 2'-DEOXY-2'-FLUOROGUANYLYL-3',5'-CYTIDINE | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE MS | | Authors: | Nonaka, T, Mitsui, Y, Nakamura, K.T. | | Deposit date: | 1991-12-02 | | Release date: | 1992-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ribonuclease Ms (as a ribonuclease T1 homologue) complexed with a guanylyl-3',5'-cytidine analogue.
Biochemistry, 32, 1993
|
|
3WO7
 
 | | Crystal structure of YidC from Bacillus halodurans (form II) | | Descriptor: | COPPER (II) ION, Membrane protein insertase YidC 2 | | Authors: | Kumazaki, K, Tsukazaki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural basis of Sec-independent membrane protein insertion by YidC.
Nature, 509, 2014
|
|
3WO6
 
 | | Crystal structure of YidC from Bacillus halodurans (form I) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CADMIUM ION, Membrane protein insertase YidC 2 | | Authors: | Kumazaki, K, Tsukazaki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Structural basis of Sec-independent membrane protein insertion by YidC.
Nature, 509, 2014
|
|
6PNN
 
 | |
4EN8
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-6-sialyllactose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
