7JKS
 
 | | Crystal structure of vaccine-elicited broadly neutralizing VRC01-class antibody 2411a in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 gp120 core, The heavy chain of antibody 2411a, ... | | Authors: | Zhou, T, Chen, X, Kwong, P.D, Mascola, J.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth.
Immunity, 54, 2021
|
|
7JKT
 
 | | Crystal structure of vaccine-elicited broadly neutralizing VRC01-class antibody 2413a in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 gp120 core from strain d45-01dG5, ... | | Authors: | Zhou, T, Chen, X, Kwong, P.D, Mascola, J.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth.
Immunity, 54, 2021
|
|
7JZ2
 
 | | Succinate: quinone oxidoreductase SQR from E.coli K12 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Lyu, M, Su, C.-C, Morgan, C.E, Bolla, J.R, Robinson, C.V, Yu, E.W. | | Deposit date: | 2020-09-01 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
7JJO
 
 | | Structural Basis of the Activation of Heterotrimeric Gs-protein by Isoproterenol-bound Beta1-Adrenergic Receptor | | Descriptor: | Beta1-Adrenergic Receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Su, M, Zhu, L, Zhang, Y, Paknejad, N, Dey, R, Huang, J, Lee, M.Y, Williams, D, Jordan, K.D, Eng, E.T, Ernst, O.P, Meyerson, J.R, Hite, R.K, Walz, T, Liu, W, Huang, X.Y. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural Basis of the Activation of Heterotrimeric Gs-Protein by Isoproterenol-Bound beta 1 -Adrenergic Receptor.
Mol.Cell, 80, 2020
|
|
7JZ3
 
 | | Osmoporin OmpC from E.coli K12 | | Descriptor: | Outer membrane protein C | | Authors: | Lyu, M, Su, C, Morgan, C.E, Bolla, J.R, Robinson, C.V, Yu, E.W. | | Deposit date: | 2020-09-01 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
3N3K
 
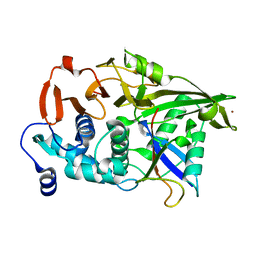 | | The catalytic domain of USP8 in complex with a USP8 specific inhibitor | | Descriptor: | Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 8, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Allali-Hassani, A, Lam, R, Ernst, A, Sidhu, S, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
7KD1
 
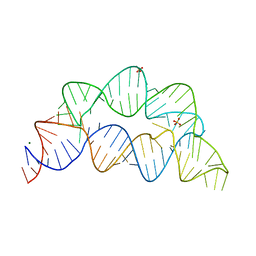 | |
7K7H
 
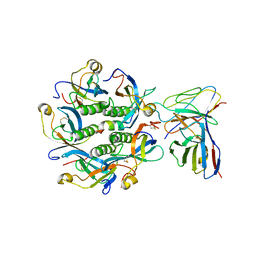 | | Density-fitted Model Structure of Antibody Variable Domains of TyTx1 in Complex with PltB pentamer of Typhoid Toxin | | Descriptor: | Fab Heavy Chain Variable Domain, Fab Light Chain Variable Domain, Pertussis like toxin subunit B, ... | | Authors: | Nguyen, T, Feathers, J.R, Fromme, J.C, Song, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The structural basis of Salmonella A 2 B 5 toxin neutralization by antibodies targeting the glycan-receptor binding subunits.
Cell Rep, 36, 2021
|
|
7K7I
 
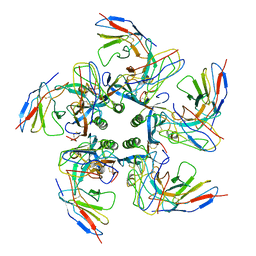 | | Density-fitted Model Structure of Antibody Variable Domains of TyTx4 in Complex with PltB pentamer of Typhoid Toxin | | Descriptor: | Fab Heavy Chain Variable Domain, Fab Light Chain Variable Domain, Putative pertussis-like toxin subunit | | Authors: | Nguyen, T, Feathers, J.R, Fromme, J.C, Song, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | The structural basis of Salmonella A 2 B 5 toxin neutralization by antibodies targeting the glycan-receptor binding subunits.
Cell Rep, 36, 2021
|
|
7KEI
 
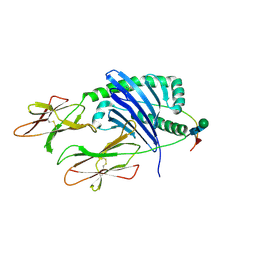 | | DQA1*01:02/DQB1*06:02 in complex with a hemagglutinin peptide from the H1N1 pandemic flu virus. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HA peptide from 2009 H1N1 pandemic flu virus., ... | | Authors: | Birtley, J.R, Stern, L.J, Mellins, E.D, Jiang, W. | | Deposit date: | 2020-10-10 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of DQA1*01:02/DQB1*06:02 in complex with a flu peptide.
To Be Published
|
|
7KTX
 
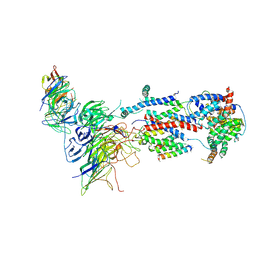 | | Cryo-EM structure of Saccharomyces cerevisiae ER membrane protein complex bound to a Fab in DDM detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 2, ... | | Authors: | Miller-Vedam, L.E, Schirle Oakdale, N.S, Braeuning, B, Boydston, E.A, Sevillano, N, Popova, K.D, Bonnar, J.L, Shurtleff, M.J, Prabu, J.R, Stroud, R.M, Craik, C.S, Schulman, B.A, Weissman, J.S, Frost, A. | | Deposit date: | 2020-11-24 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
7KRA
 
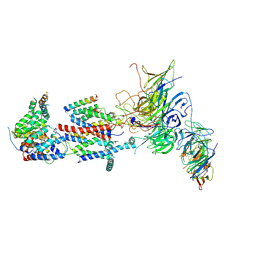 | | Cryo-EM structure of Saccharomyces cerevisiae ER membrane protein complex bound to Fab-DH4 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 2, ... | | Authors: | Miller-Vedam, L.E, Schirle Oakdale, N.S, Braeuning, B, Boydston, E.A, Sevillano, N, Popova, K.D, Bonnar, J.L, Shurtleff, M.J, Prabu, J.R, Stroud, R.M, Craik, C.S, Schulman, B.A, Weissman, J.S, Frost, A. | | Deposit date: | 2020-11-19 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
10MH
 
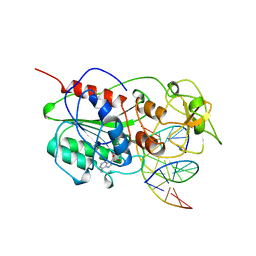 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH ADOHCY AND HEMIMETHYLATED DNA CONTAINING 5,6-DIHYDRO-5-AZACYTOSINE AT THE TARGET | | Descriptor: | DNA (5'-D(P*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*AP*GP*5NCP*GP*CP*AP*TP*GP*G)-3'), PROTEIN (CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI), ... | | Authors: | Sheikhnejad, G, Brank, A, Christman, J.K, Goddard, A, Alvarez, E, Ford Junior, H, Marquez, V.E, Marasco, C.J, Sufrin, J.R, O'Gara, M, Cheng, X. | | Deposit date: | 1998-08-10 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanism of inhibition of DNA (cytosine C5)-methyltransferases by oligodeoxyribonucleotides containing 5,6-dihydro-5-azacytosine.
J.Mol.Biol., 285, 1999
|
|
1AH5
 
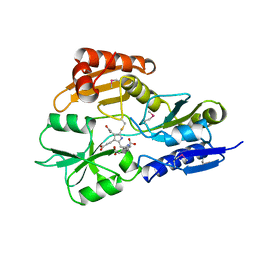 | | REDUCED FORM SELENOMETHIONINE-LABELLED HYDROXYMETHYLBILANE SYNTHASE DETERMINED BY MAD | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, HYDROXYMETHYLBILANE SYNTHASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Harrop, S.J, Cassetta, A. | | Deposit date: | 1997-04-13 | | Release date: | 1997-10-15 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of the structure of seleno-methionine-labelled hydroxymethylbilane synthase in its active form by multi-wavelength anomalous dispersion.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1IKI
 
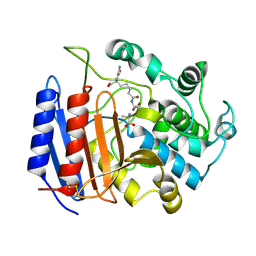 | | COMPLEX OF STREPTOMYCES R61 DD-PEPTIDASE WITH THE PRODUCTS OF A SPECIFIC PEPTIDOGLYCAN SUBSTRATE FRAGMENT | | Descriptor: | D-ALANINE, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, GLYCYL-L-ALPHA-AMINO-EPSILON-PIMELYL-D-ALANINE | | Authors: | Mcdonough, M.A, Anderson, J.W, Silvaggi, N.R, Pratt, R.F, Knox, J.R, Kelly, J.A. | | Deposit date: | 2001-05-03 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of two kinetic intermediates reveal species specificity of penicillin-binding proteins.
J.Mol.Biol., 322, 2002
|
|
184D
 
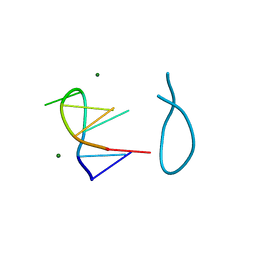 | | SELF-ASSOCIATION OF A DNA LOOP CREATES A QUADRUPLEX: CRYSTAL STRUCTURE OF D(GCATGCT) AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3'), MAGNESIUM ION | | Authors: | Leonard, G.A, Zhang, S, Peterson, M.R, Harrop, S.J, Helliwell, J.R, Cruse, W.B.T, Langlois D'Estaintot, B, Kennard, O, Brown, T, Hunter, W.N. | | Deposit date: | 1994-08-10 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Self-association of a DNA loop creates a quadruplex: crystal structure of d(GCATGCT) at 1.8 A resolution.
Structure, 3, 1995
|
|
1AH2
 
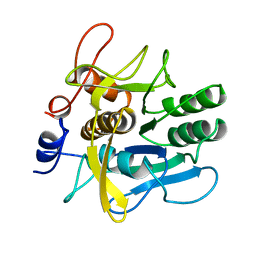 | | SERINE PROTEASE PB92 FROM BACILLUS ALCALOPHILUS, NMR, 18 STRUCTURES | | Descriptor: | SERINE PROTEASE PB92 | | Authors: | Boelens, R, Schipper, D, Martin, J.R, Karimi-Nejad, Y, Mulder, F, Zwan, J.V.D, Mariani, M. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of serine protease PB92 from Bacillus alcalophilus presents a rigid fold with a flexible substrate-binding site.
Structure, 5, 1997
|
|
1AVO
 
 | | PROTEASOME ACTIVATOR REG(ALPHA) | | Descriptor: | 11S REGULATOR | | Authors: | Hill, C.P, Knowlton, J.R. | | Deposit date: | 1997-09-18 | | Release date: | 1997-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the proteasome activator REGalpha (PA28alpha).
Nature, 390, 1997
|
|
1H5O
 
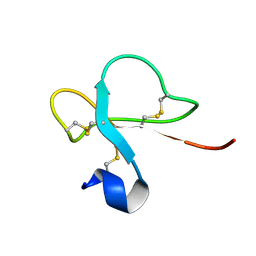 | | Solution structure of Crotamine, a neurotoxin from Crotalus durissus terrificus | | Descriptor: | MYOTOXIN | | Authors: | Nicastro, G, Franzoni, L, De Chiara, C, Mancin, C.A, Giglio, J.R, Spisni, A. | | Deposit date: | 2001-05-23 | | Release date: | 2003-05-09 | | Last modified: | 2013-07-24 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Crotamine, a Na+ Channel Affecting Toxin from Crotalus Durissus Terrificus Venom
Eur.J.Biochem., 270, 2003
|
|
1GTK
 
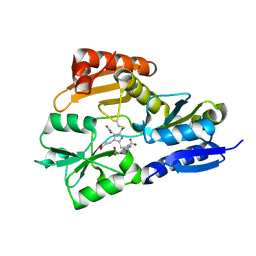 | | Time-resolved and static-ensemble structural chemistry of hydroxymethylbilane synthase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, PORPHOBILINOGEN DEAMINASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Raftery, J, Cassetta, A, Habash, J, Carr, P.D, Ursby, T, Wulff, M, Thompson, A.W, Niemann, A.C, Haedener, A. | | Deposit date: | 2002-01-16 | | Release date: | 2003-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Time-Resolved and Static-Ensemble Structural Chemistry of Hydroxymethylbilane Synthase
Faraday Discuss., 122, 2003
|
|
1H0R
 
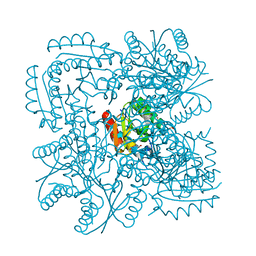 | | Type II Dehydroquinase from Mycobacterium tuberculosis complexed with 2,3-anhydro-quinic acid | | Descriptor: | 2,3 -ANHYDRO-QUINIC ACID, 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | Roszak, A.W, Robinson, D.A, Frederickson, M, Abell, C, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-06-27 | | Release date: | 2003-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Selectivity of Oxime Based Inhibitors Towards Type II Dehydroquinase from Mycobacterium Tuberculosis
To be Published
|
|
1JJG
 
 | | Solution Structure of Myxoma Virus Protein M156R | | Descriptor: | M156R | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-07-05 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Myxoma virus immunomodulatory protein M156R is a structural mimic of eukaryotic translation initiation factor eIF2alpha.
J.Mol.Biol., 322, 2002
|
|
2FN2
 
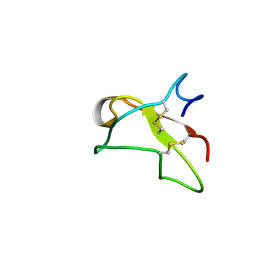 | | SOLUTION NMR STRUCTURE OF THE GLYCOSYLATED SECOND TYPE TWO MODULE OF FIBRONECTIN, 20 STRUCTURES | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FIBRONECTIN | | Authors: | Sticht, H, Pickford, A.R, Potts, J.R, Campbell, I.D. | | Deposit date: | 1997-08-06 | | Release date: | 1998-09-16 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the glycosylated second type 2 module of fibronectin.
J.Mol.Biol., 276, 1998
|
|
1AIU
 
 | | HUMAN THIOREDOXIN (D60N MUTANT, REDUCED FORM) | | Descriptor: | THIOREDOXIN | | Authors: | Andersen, J.F, Gasdaska, J.R, Sanders, D.A.R, Weichsel, A, Powis, G, Montfort, W.R. | | Deposit date: | 1997-04-25 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human thioredoxin homodimers: regulation by pH, role of aspartate 60, and crystal structure of the aspartate 60 --> asparagine mutant.
Biochemistry, 36, 1997
|
|
1A8A
 
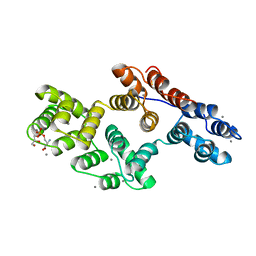 | | RAT ANNEXIN V COMPLEXED WITH GLYCEROPHOSPHOSERINE | | Descriptor: | ANNEXIN V, CALCIUM ION, L-ALPHA-GLYCEROPHOSPHORYLSERINE | | Authors: | Swairjo, M.A, Concha, N.O, Kaetzel, M.A, Dedman, J.R, Seaton, B.A. | | Deposit date: | 1998-03-23 | | Release date: | 1998-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ca(2+)-bridging mechanism and phospholipid head group recognition in the membrane-binding protein annexin V.
Nat.Struct.Biol., 2, 1995
|
|
