5MAG
 
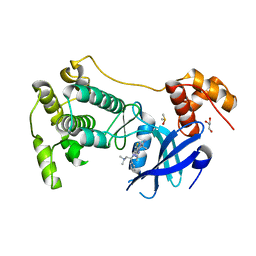 | | Crystal structure of MELK in complex with an inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Maternal embryonic leucine zipper kinase, ... | | Authors: | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
6RDK
 
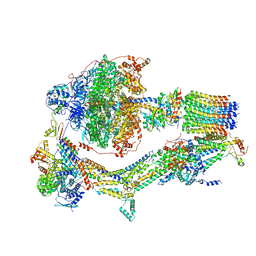 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1B, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RE0
 
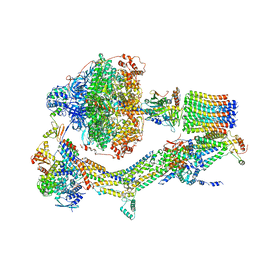 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2A, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6REP
 
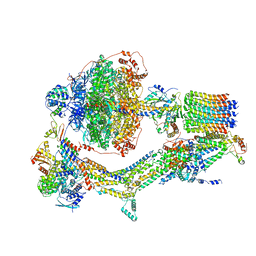 | | Cryo-EM structure of Polytomella F-ATP synthase, Primary rotary state 3, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
5JFW
 
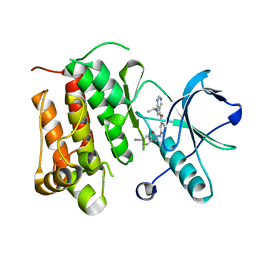 | | Crystal structure of TrkA in complex with PF-05247452 | | Descriptor: | 2-(4-cyanophenyl)-N-{5-[7-(propan-2-yl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonyl]pyridin-3-yl}acetamide, High affinity nerve growth factor receptor | | Authors: | Jayasankar, J, Kurumbail, R, Skerratt, S, Brown, D. | | Deposit date: | 2016-04-19 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The Discovery of a Potent, Selective, and Peripherally Restricted Pan-Trk Inhibitor (PF-06273340) for the Treatment of Pain.
J. Med. Chem., 59, 2016
|
|
6RD9
 
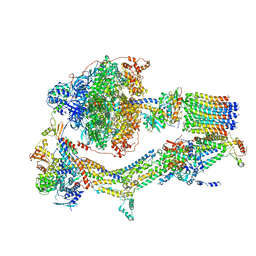 | | CryoEM structure of Polytomella F-ATP synthase, Primary rotary state 1, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDT
 
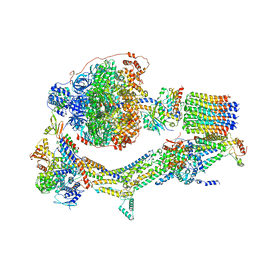 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1E, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6REA
 
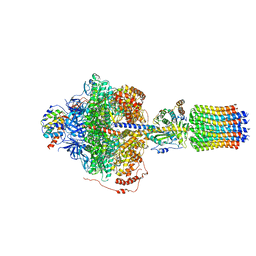 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2D, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RES
 
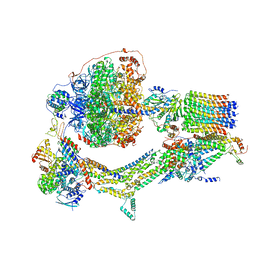 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3C, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6DG6
 
 | | Structure of a de novo designed Interleukin-2/Interleukin-15 mimetic | | Descriptor: | Neoleukin-2/15 | | Authors: | Jude, K.M, Silva, D.-A, Yu, S, Baker, D, Garcia, K.C. | | Deposit date: | 2018-05-16 | | Release date: | 2019-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | De novo design of potent and selective mimics of IL-2 and IL-15.
Nature, 565, 2019
|
|
5IGR
 
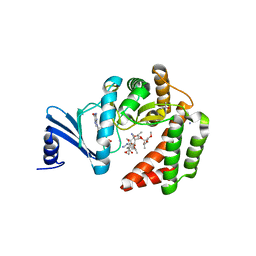 | | Macrolide 2'-phosphotransferase type I - complex with GDP and oleandomycin | | Descriptor: | (3S,5R,6S,7R,8R,11R,12S,13R,14S,15S)-6-HYDROXY-5,7,8,11,13,15-HEXAMETHYL-4,10-DIOXO-14-{[3,4,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-XYLO-HEXOPYRANOSYL]OXY}-1,9-DIOXASPIRO[2.13]HEXADEC-12-YL 2,6-DIDEOXY-3-O-METHYL-ALPHA-L-ARABINO-HEXOPYRANOSIDE, AMMONIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Berghuis, A.M, Fong, D.H. | | Deposit date: | 2016-02-28 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Kinase-Mediated Macrolide Antibiotic Resistance.
Structure, 25, 2017
|
|
5IH1
 
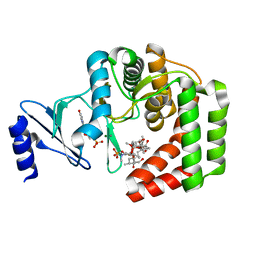 | |
5MAH
 
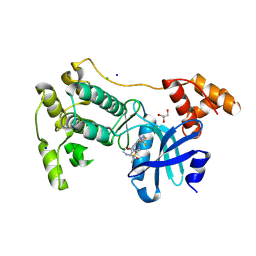 | | Crystal structure of MELK in complex with an inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, Maternal embryonic leucine zipper kinase, ... | | Authors: | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5IGU
 
 | | Macrolide 2'-phosphotransferase type II | | Descriptor: | Macrolide 2'-phosphotransferase II | | Authors: | Berghuis, A.M, Fong, D.H. | | Deposit date: | 2016-02-28 | | Release date: | 2017-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Kinase-Mediated Macrolide Antibiotic Resistance.
Structure, 25, 2017
|
|
5IH0
 
 | |
5MAI
 
 | | Crystal structure of MELK in complex with an inhibitor | | Descriptor: | 3-[(3~{Z})-3-[[[4-[(dimethylamino)methyl]phenyl]amino]-phenyl-methylidene]-2-oxidanylidene-1~{H}-indol-6-yl]-~{N}-ethyl-prop-2-ynamide, DIMETHYL SULFOXIDE, Maternal embryonic leucine zipper kinase | | Authors: | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
7K9Z
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with the Fab fragments of neutralizing antibodies 298 and 52 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 298 Fab Heavy Chain, 298 Fab Light Chain, ... | | Authors: | Newton, J.C, Kucharska, I, Rujas, E, Cui, H, Julien, J.P. | | Deposit date: | 2020-09-29 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Multivalency transforms SARS-CoV-2 antibodies into ultrapotent neutralizers.
Nat Commun, 12, 2021
|
|
5ITD
 
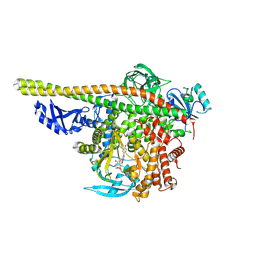 | | Crystal structure of PI3K alpha with PI3K delta inhibitor | | Descriptor: | 5-{4-[3-(4-acetylpiperazine-1-carbonyl)phenyl]quinazolin-6-yl}-2-methoxypyridine-3-carbonitrile, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2016-03-16 | | Release date: | 2016-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Discovery and Pharmacological Characterization of Novel Quinazoline-Based PI3K Delta-Selective Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5IGJ
 
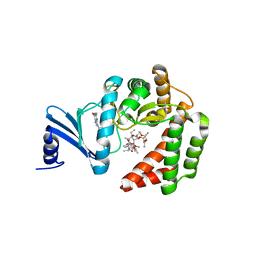 | |
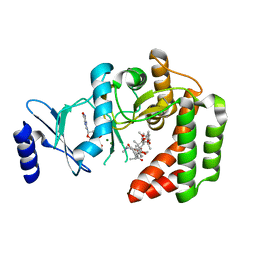
5IGW
 
 | |
4KZ5
 
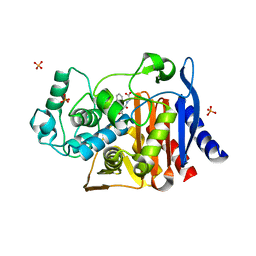 | | Crystal structure of AmpC beta-lactamase in complex with fragment 5 (N-{[3-(2-chlorophenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl}glycine) | | Descriptor: | Beta-lactamase, N-{[3-(2-chlorophenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl}glycine, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
5IGI
 
 | |
5IGT
 
 | |
5D1Z
 
 | | IsdB NEAT1 bound by clone D4-10 | | Descriptor: | D4-10 Heavy Chain, D4-10 Light Chain, Iron-regulated surface determinant protein B, ... | | Authors: | Deng, X. | | Deposit date: | 2015-08-04 | | Release date: | 2016-09-14 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Germline-encoded neutralization of a Staphylococcus aureus virulence factor by the human antibody repertoire.
Nat Commun, 7, 2016
|
|
4Y73
 
 | | Crystal structure of IRAK4 kinase domain with inhibitor | | Descriptor: | 5-{[(1R,2S)-2-aminocyclohexyl]amino}-N-[1-methyl-3-(trifluoromethyl)-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Lesburg, C.A. | | Deposit date: | 2015-02-13 | | Release date: | 2015-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of 5-Amino-N-(1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide Inhibitors of IRAK4.
Acs Med.Chem.Lett., 6, 2015
|
|
