6WL0
 
 | | Cryo-EM of Form 1 related peptide filament, 36-31-3-RD | | Descriptor: | peptide 36-31-3-RD | | Authors: | Wang, F, Gnewou, O.M, Su, Z, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
6WL9
 
 | | Cryo-EM of Form 2 like peptide filament, Form2a | | Descriptor: | peptide Form2a | | Authors: | Wang, F, Beltran, L.C, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
6WL8
 
 | | Cryo-EM of Form 2 peptide filament | | Descriptor: | Form 2 peptide | | Authors: | Wang, F, Gnewou, O.M, Xu, C, Su, Z, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
5LV0
 
 | | Structure of Human Neurolysin (E475Q) in complex with amyloid-beta 35-40 peptide product | | Descriptor: | CHLORIDE ION, GLY-VAL-VAL amyloid 35-40 fragment, Neurolysin, ... | | Authors: | Masuyer, G, Berntsson, R.P.-A, Teixeira, P.F, Kmiec, B, Glaser, E, Stenmark, P. | | Deposit date: | 2016-09-12 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional analysis of Neurolysin, a new component of the mitochondrial peptidolytic network
To Be Published
|
|
7ASJ
 
 | |
7BTW
 
 | | The mitochondrial SAM complex from S.cere | | Descriptor: | Mitochondrial outer membrane beta-barrel protein, SAM37 isoform 1, Sorting assembly machinery 35 kDa subunit | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
7BTX
 
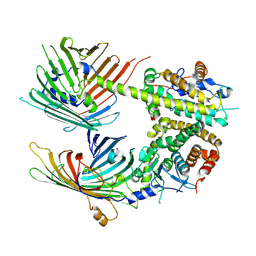 | | The mitochondrial SAM-Mdm10 supercomplex in GDN micelle from S.cere | | Descriptor: | MDM10 isoform 1, Mitochondrial outer membrane beta-barrel protein, Sorting assembly machinery 35 kDa subunit, ... | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
7BTY
 
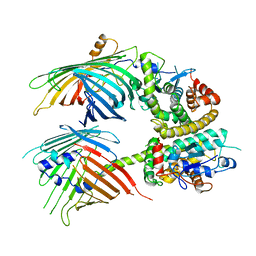 | | The mitochondrial SAM-Mdm10 supercomplex in Nanodisc from S.cere | | Descriptor: | MDM10 isoform 1, Mitochondrial outer membrane beta-barrel protein, Sorting assembly machinery 35 kDa subunit, ... | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
5OGB
 
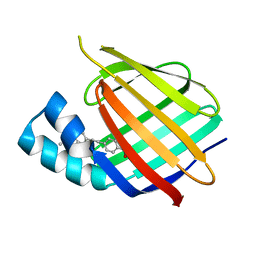 | | Human Cellular Retinoic Acid Binding Protein II (CRABPII) with bound synthetic retinoid DC360. | | Descriptor: | 4-[2-(4,4-dimethyl-1-propan-2-yl-quinolin-6-yl)ethynyl]benzoic acid, Cellular retinoic acid-binding protein 2 | | Authors: | Chisholm, D, Tomlinson, C, Whiting, A, Pohl, E. | | Deposit date: | 2017-07-12 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fluorescent Retinoic Acid Analogues as Probes for Biochemical and Intracellular Characterization of Retinoid Signaling Pathways.
Acs Chem.Biol., 14, 2019
|
|
7PH1
 
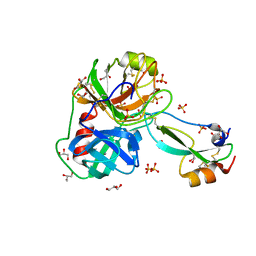 | | Trypsin in complex with BPTI mutant (2S)-2-amino-4-monofluorobutanoic acid | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Loll, B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Water Network in the Binding Pocket of Fluorinated BPTI-Trypsin Complexes─Insights from Simulation and Experiment.
J.Phys.Chem.B, 126, 2022
|
|
6HM7
 
 | |
6KEU
 
 | | Wildtype E53, a microbial HSL esterase | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Yang, X.C, Li, Z.Y, Xu, X.W, Li, J.X. | | Deposit date: | 2019-07-05 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Wildtype E53, a microbial HSL esterase
To Be Published
|
|
6HM6
 
 | |
6KF1
 
 | | Microbial Hormone-sensitive lipase- E53 mutant S162A | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HEXANOIC ACID, ... | | Authors: | Xiaochen, Y, Zhengyang, L, Xuewei, X, Jixi, L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Functional and Structural Insights into Environmental Adaptation of a Novel Hormone-sensitive Lipase, E53, Obtained from Erythrobacter longus
To Be Published
|
|
8ORC
 
 | | Mus Musculus Acetylcholinesterase in complex with AL237 | | Descriptor: | 1-[2-(dimethylamino)ethyl]-3-(2-methoxyphenyl)thiourea, 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Ekstrom, F.E, Linusson, A. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enzyme Dynamics Determine the Potency and Selectivity of Inhibitors Targeting Disease-Transmitting Mosquitoes.
Acs Infect Dis., 10, 2024
|
|
6JNF
 
 | | Cryo-EM structure of the translocator of the outer mitochondrial membrane | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Araiso, Y, Tsutsumi, A, Suzuki, J, Yunoki, K, Kawano, S, Kikkawa, M, Endo, T. | | Deposit date: | 2019-03-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structure of the mitochondrial import gate reveals distinct preprotein paths.
Nature, 575, 2019
|
|
6I8F
 
 | |
5LUZ
 
 | | Structure of Human Neurolysin (E475Q) in complex with neurotensin peptide products | | Descriptor: | CHLORIDE ION, GLYCEROL, Neurolysin, ... | | Authors: | Masuyer, G, Berntsson, R.P.-A, Teixeira, P.F, Kmiec, B, Glaser, E, Stenmark, P. | | Deposit date: | 2016-09-12 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Peptide Binding and Cleavage by the Human Mitochondrial Peptidase Neurolysin.
J. Mol. Biol., 430, 2018
|
|
6BFB
 
 | | Crystal structure of a F. nucleatum FMN riboswitch bound to WG-3 | | Descriptor: | 5-[(3S,4S)-3-(dimethylamino)-4-hydroxypyrrolidin-1-yl]-6-fluoro-4-methyl-8-oxo-3,4-dihydro-8H-1-thia-4,9b-diazacyclopenta[cd]phenalene-9-carboxylic acid, MAGNESIUM ION, RNA (54-MER), ... | | Authors: | Rizvi, N.F, Fischmann, T.O. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery of Selective RNA-Binding Small Molecules by Affinity-Selection Mass Spectrometry.
ACS Chem. Biol., 13, 2018
|
|
6B4D
 
 | | Crystal structure of human carbonic anhydrase II in complex with a heteroaryl-pyrazole carboxylic acid derivative. | | Descriptor: | 3-(1-ethyl-1H-indol-3-yl)-1-methyl-1H-pyrazole-5-carboxylic acid, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Lomelino, C.L, Mahon, B.P, McKenna, R. | | Deposit date: | 2017-09-26 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.196 Å) | | Cite: | Exploring Heteroaryl-pyrazole Carboxylic Acids as Human Carbonic Anhydrase XII Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
6RKY
 
 | |
7ZR3
 
 | |
1N4H
 
 | | Characterization of ligands for the orphan nuclear receptor RORbeta | | Descriptor: | Nuclear Receptor ROR-beta, RETINOIC ACID, Steroid Receptor Coactivator-1 | | Authors: | Stehlin-Gaon, C, Willmann, D, Sanglier, S, Van Dorsselaer, A, Renaud, J.-P, Moras, D, Schuele, R. | | Deposit date: | 2002-10-31 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | All-trans retinoic acid is a ligand for the orphan nuclear receptor RORbeta
Nat.Struct.Biol., 10, 2003
|
|
1DPI
 
 | |
8AYV
 
 | |
