8G92
 
 | | Structure of inhibitor 16d-bound SPNS2 | | Descriptor: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Chen, H, Li, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
8YN5
 
 | | Cryo-EM structure of histamine H3 receptor in complex with histamine and Gi | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
5WQ7
 
 | |
5WJ9
 
 | | Human TRPML1 channel structure in agonist-bound open conformation | | Descriptor: | 2-{2-oxo-2-[(4S)-2,2,4-trimethyl-3,4-dihydroquinolin-1(2H)-yl]ethyl}-1H-isoindole-1,3(2H)-dione, Mucolipin-1 | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2017-07-21 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Human TRPML1 channel structures in open and closed conformations.
Nature, 550, 2017
|
|
7F7I
 
 | | Stapled Peptide Inhibitor in complex with PSD95 GK domain | | Descriptor: | ACE-ARG-ILE-ARG-ARG-ASP-GLU-TYR-LEU-LYZ-ALA-ILE-GLN-NH2, Disks large homolog 4 | | Authors: | Shang, Y, Huang, X, Li, X, Zhang, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.
Rsc Chem Biol, 2, 2021
|
|
5WQ8
 
 | |
8YNA
 
 | | Cryo-EM structure of histamine H4 receptor in complex with immepip and Gi | | Descriptor: | 4-(1H-imidazol-5-ylmethyl)piperidine, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN9
 
 | | Cryo-EM structure of histamine H4 receptor in complex with histamine and Gi | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN2
 
 | | Cryo-EM structure of histamine H1 receptor in complex with histamine and miniGq | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Engineered guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN4
 
 | | Cryo-EM structure of histamine H2 receptor in complex with histamine and miniGq | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Engineered guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN6
 
 | | Cryo-EM structure of histamine H3 receptor in complex with imetit and Gi | | Descriptor: | 2-(1~{H}-imidazol-5-yl)ethyl carbamimidothioate, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
5XRA
 
 | | Crystal structure of the human CB1 in complex with agonist AM11542 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6aR,10aR)-3-(8-bromanyl-2-methyl-octan-2-yl)-6,6,9-trimethyl-6a,7,10,10a-tetrahydrobenzo[c]chromen-1-ol, CHOLESTEROL, ... | | Authors: | Hua, T, Vemuri, K, Nikas, P.S, Laprairie, R.B, Wu, Y, Qu, L, Pu, M, Korde, A, Shan, J, Ho, J.H, Han, G.W, Ding, K, Li, X, Liu, H, Hanson, M.A, Zhao, S, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of agonist-bound human cannabinoid receptor CB1
Nature, 547, 2017
|
|
8YN3
 
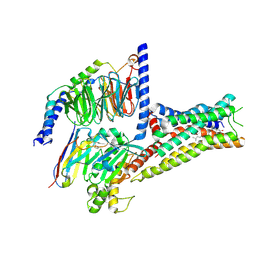 | | Cryo-EM structure of histamine H2 receptor in complex with histamine and miniGs | | Descriptor: | CHOLESTEROL, Engineered guanine nucleotide,binding protein G(s) subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN8
 
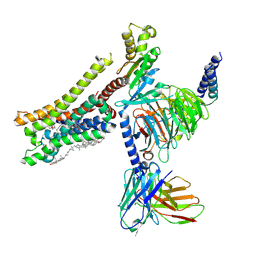 | | Cryo-EM structure of histamine H3 receptor in complex with proxyfan and miniGo | | Descriptor: | 5-(3-phenylmethoxypropyl)-1H-imidazole, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN7
 
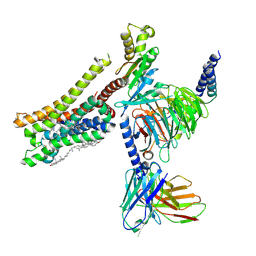 | | Cryo-EM structure of histamine H3 receptor in complex with immethridine and miniGo | | Descriptor: | 4-(1H-imidazol-4-ylmethyl)pyridine, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
5WQ9
 
 | |
5WJ5
 
 | |
5V93
 
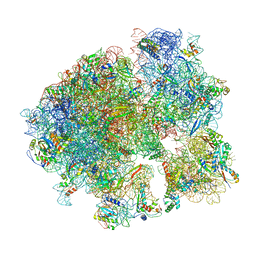 | | Cryo-EM structure of the 70S ribosome from Mycobacterium tuberculosis bound with Capreomycin | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Yang, K, Chang, J.-Y, Cui, Z, Li, X, Meng, R, Duan, L, Thongchol, J, Jakana, J, Huwe, C, Sacchettini, J, Zhang, J. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-20 | | Last modified: | 2020-08-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis.
Nucleic Acids Res., 45, 2017
|
|
7KHR
 
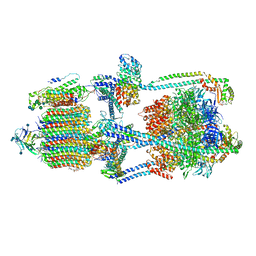 | | Cryo-EM structure of bafilomycin A1-bound intact V-ATPase from bovine brain | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (5R)-2,4-dideoxy-1-C-{(2S,3R,4S)-3-hydroxy-4-[(2R,3S,4E,6E,9R,10S,11R,12E,14Z)-10-hydroxy-3,15-dimethoxy-7,9,11,13-tetramethyl-16-oxo-1-oxacyclohexadeca-4,6,12,14-tetraen-2-yl]pentan-2-yl}-4-methyl-5-propan-2-yl-alpha-D-threo-pentopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Molecular basis of V-ATPase inhibition by bafilomycin A1.
Nat Commun, 12, 2021
|
|
5XR8
 
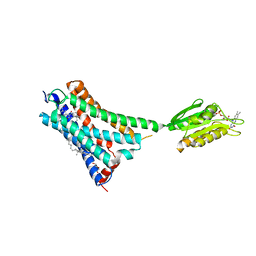 | | Crystal structure of the human CB1 in complex with agonist AM841 | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, CHOLESTEROL, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Nikas, P.S, Laprairie, R.B, Wu, Y, Qu, L, Pu, M, Korde, A, Shan, J, Ho, J.H, Han, G.W, Ding, K, Li, X, Liu, H, Hanson, M.A, Zhao, S, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of agonist-bound human cannabinoid receptor CB1.
Nature, 547, 2017
|
|
6JIR
 
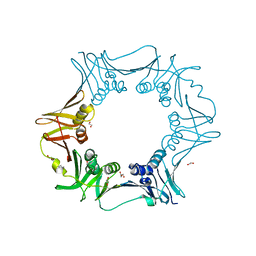 | | Crystal structure of C. crescentus beta sliding clamp with PEG bound to putative beta-motif tethering region | | Descriptor: | 1,2-ETHANEDIOL, Beta sliding clamp, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jiang, X, Teng, M, Li, X. | | Deposit date: | 2019-02-23 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Caulobacter crescentus beta sliding clamp employs a noncanonical regulatory model of DNA replication.
Febs J., 287, 2020
|
|
6KJ3
 
 | | 120kV MicroED structure of FUS (37-42) SYSGYS solved from merged datasets at 0.60 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.6 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6KG7
 
 | | Cryo-EM Structure of the Mammalian Tactile Channel Piezo2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Piezo-type mechanosensitive ion channel component 2 | | Authors: | Wang, L, Zhou, H, Zhang, M, Liu, W, Deng, T, Zhao, Q, Li, Y, Lei, J, Li, X, Xiao, B. | | Deposit date: | 2019-07-11 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and mechanogating of the mammalian tactile channel PIEZO2.
Nature, 573, 2019
|
|
6KJ4
 
 | | 120kV MicroED structure of FUS (37-42) SYSGYS solved from single crystal at 0.65 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.65 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6KJ2
 
 | | 200kV MicroED structure of FUS (37-42) SYSGYS solved from single crystal at 0.67 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.67 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
