9BG1
 
 | | Tri-complex of Compound-3, KRAS G12V, and CypA | | Descriptor: | (2R)-N-[(1P,7S,9S,13R,20M)-21-ethyl-20-{2-[(1S)-1-methoxyethyl]pyridin-3-yl}-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-methyl-2-(N-methylacetamido)butanamide (non-preferred name), GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-04-18 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Tri-complex of Compound-3, KRAS G12V, and CypA
To be published
|
|
5EHR
 
 | |
5EHP
 
 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor SHP836 | | Descriptor: | 5-[2,3-bis(chloranyl)phenyl]-2-[(3~{R},5~{S})-3,5-dimethylpiperazin-1-yl]pyrimidin-4-amine, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2015-10-28 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Allosteric Inhibition of SHP2: Identification of a Potent, Selective, and Orally Efficacious Phosphatase Inhibitor.
J.Med.Chem., 59, 2016
|
|
6LGN
 
 | | The atomic structure of varicella zoster virus C-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Li, S, Zheng, Q. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
7WY5
 
 | | ADGRL3/Gq complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling.
Mol.Cell, 82, 2022
|
|
7WY8
 
 | | ADGRL3/Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling
Mol.Cell, 82, 2022
|
|
7WYB
 
 | | ADGRL3/Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and G q , G s , G i , and G 12 coupling.
Mol.Cell, 82, 2022
|
|
7X10
 
 | | ADGRL3/miniG12 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-22 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling.
Mol.Cell, 82, 2022
|
|
4LYR
 
 | | Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E301A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Exo-beta-1,4-mannosidase | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LYQ
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E202A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Exo-beta-1,4-mannosidase, ... | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8JIO
 
 | |
9C1P
 
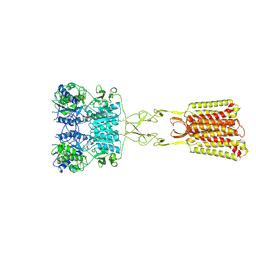 | | Structure of Calcium-Sensing Receptor in complex with positive allosteric modulator '6218 | | Descriptor: | (5R)-N-[2-(1,2-benzothiazol-3-yl)ethyl]-1-methyl-2,3,4,5-tetrahydro-1H-1-benzazepin-5-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, C, Skiniotis, G. | | Deposit date: | 2024-05-29 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Large library docking identifies positive allosteric modulators of the calcium-sensing receptor.
Science, 385, 2024
|
|
9C2F
 
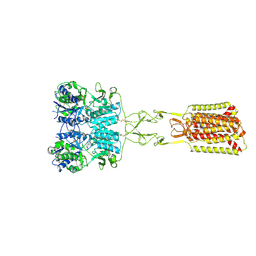 | | Structure of Calcium-Sensing Receptor in complex with positive allosteric modulator '54149 | | Descriptor: | (1R)-1-(2H-1,3-benzodioxol-4-yl)-N-[2-(1,2-benzothiazol-3-yl)ethyl]ethan-1-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, C, Skiniotis, G. | | Deposit date: | 2024-05-30 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Large library docking identifies positive allosteric modulators of the calcium-sensing receptor.
Science, 385, 2024
|
|
7XNF
 
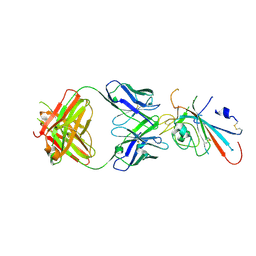 | | Structure of SARS-CoV-2 antibody P2C-1F11 with GX/P2V/2017 RBD | | Descriptor: | P2C-1F11 Heavy Chain, P2C-1F11 Lambda chain, Spike protein S1 | | Authors: | Jia, Y.F, Chai, Y, Wang, Q.H, Gao, G.F. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Cross-reaction of current available SARS-CoV-2 MAbs against the pangolin-origin coronavirus GX/P2V/2017.
Cell Rep, 41, 2022
|
|
7XBF
 
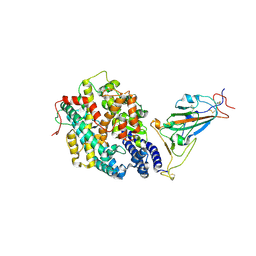 | | The complex structure of RshSTT182/200 RBD-insert2 bound to human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, RshSTT182/200 coronavirus receptor binding domain insert2, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
7XBG
 
 | | The crystal structure of RshSTT182/200 RBD-insert2-T346R-Y496G mutant in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
7XBH
 
 | | The complex structure of RshSTT182/200 RBD bound to human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, RshSTT182/200 coronavirus receptor binding domain, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
3BXV
 
 | |
8J54
 
 | | Crystal structure of RXR/DR2 complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*CP*CP*TP*AP*CP*TP*GP*AP*CP*CP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*GP*GP*TP*CP*AP*GP*TP*AP*GP*GP*TP*CP*AP*TP*G)-3'), Retinoic acid receptor RXR, ... | | Authors: | Chen, Y, Jiang, L. | | Deposit date: | 2023-04-21 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural characterization of the DNA binding mechanism of retinoic acid-related orphan receptor gamma.
Structure, 32, 2024
|
|
4DZB
 
 | |
7X4X
 
 | | BTB domain of KEAP1 in complex with MEF | | Descriptor: | 4-ethoxy-4-oxobutanoic acid, Kelch-like ECH-associated protein 1 | | Authors: | Qu, L.Z. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Characterization of the modification of Kelch-like ECH-associated protein 1 by different fumarates.
Biochem.Biophys.Res.Commun., 605, 2022
|
|
7X4W
 
 | |
7XVN
 
 | | Structural basis for DNA recognition feature of retinoid-related orphan receptors | | Descriptor: | DNA (5'-D(P*CP*AP*TP*GP*AP*CP*CP*TP*AP*CP*TP*GP*AP*CP*CP*TP*AP*G)-3'), DNA (5'-D(P*CP*TP*AP*GP*GP*TP*CP*AP*GP*TP*AP*GP*GP*TP*CP*AP*TP*G)-3'), Nuclear receptor ROR-gamma, ... | | Authors: | Chen, Y, Jiang, L. | | Deposit date: | 2022-05-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural characterization of the DNA binding mechanism of retinoic acid-related orphan receptor gamma.
Structure, 32, 2024
|
|
7WZ2
 
 | | SARS-CoV-2 (D614G) Spike trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-02-16 | | Release date: | 2022-07-20 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WZ1
 
 | | SARS-CoV-2 Omicron Spike trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-02-16 | | Release date: | 2022-07-27 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
