5MJL
 
 | | Single-shot pink beam serial crystallography: Proteinase K | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.21013784 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
5N74
 
 | | Microtubule end binding protein complex | | Descriptor: | Karyogamy protein KAR9, Microtubule-associated protein RP/EB family member 1 | | Authors: | Kumar, A, Steinmetz, M. | | Deposit date: | 2017-02-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Short Linear Sequence Motif LxxPTPh Targets Diverse Proteins to Growing Microtubule Ends.
Structure, 25, 2017
|
|
5NJS
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 50s time-delay, phased with 1HEW | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
5NJP
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 1s time-delay, phased with 1HEW | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
5NJM
 
 | | Lysozyme room-temperature structure determined by serial millisecond crystallography | | Descriptor: | Lysozyme C | | Authors: | Weinert, T, Vera, L, Marsh, M, James, D, Gashi, D, Nogly, P, Jaeger, K, Standfuss, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5NJQ
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 1s time-delay, phased with 4ET8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
5NLX
 
 | | A2A Adenosine receptor room-temperature structure determined by serial millisecond crystallography | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Dore, A.S, Geng, T, Cooke, R, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5O7M
 
 | | Single-shot pink beam serial crystallography: Phycocyanin (One chip, chip_1) | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V, Sarrou, I. | | Deposit date: | 2017-06-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
5O5W
 
 | | Molybdenum storage protein room-temperature structure determined by serial millisecond crystallography | | Descriptor: | (mu3-oxo)-tris(mu2-oxo)-nonakisoxo-trimolybdenum (VI), ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Steffen, B, Weinert, T, Ermler, U, Standfuss, J. | | Deposit date: | 2017-06-02 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
4TMS
 
 | |
1GN2
 
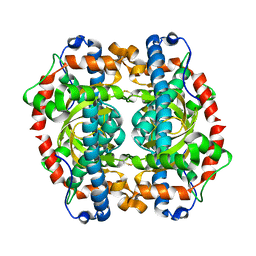 | | S123C mutant of the iron-superoxide dismutase from Mycobacterium tuberculosis. | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Bunting, K.A, Cooper, J.B, Tickle, I.J, Young, D.B. | | Deposit date: | 2001-10-02 | | Release date: | 2001-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Engineering of an Intersubunit Disulfide Bridge in the Iron-Superoxide Dismutase of Mycobacterium Tuberculosis.
Arch.Biochem.Biophys., 397, 2002
|
|
1HDS
 
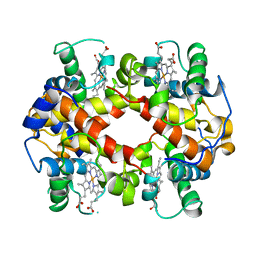 | |
2JTE
 
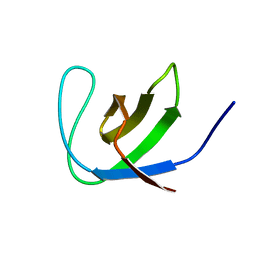 | | Third SH3 domain of CD2AP | | Descriptor: | CD2-associated protein | | Authors: | van Nuland, N.A.J, Ortega, R.J, Romero Romero, M, Ab, E, Ora, A, Lopez, M.O, Azuaga, A.I. | | Deposit date: | 2007-07-28 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The high resolution NMR structure of the third SH3 domain of CD2AP.
J.Biomol.Nmr, 39, 2007
|
|
7Z01
 
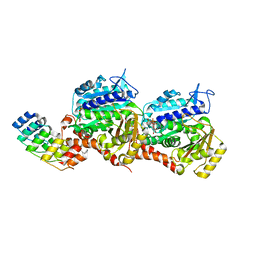 | | Z-SBTubA4 photoswitch bound to tubulin-DARPin D1 complex | | Descriptor: | 2-methoxy-5-[2-(5,6,7-trimethoxy-1,3-benzothiazol-2-yl)ethyl]phenol, CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Wranik, M, Weinert, T, Standfuss, J, Steinmetz, M. | | Deposit date: | 2022-02-21 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | In Vivo Photocontrol of Microtubule Dynamics and Integrity, Migration and Mitosis, by the Potent GFP-Imaging-Compatible Photoswitchable Reagents SBTubA4P and SBTub2M.
J.Am.Chem.Soc., 144, 2022
|
|
7Z02
 
 | | Z-SBTub2M photoswitch bound to tubulin-DARPin D1 complex | | Descriptor: | 6-methyl-2-[2-(3,4,5-trimethoxyphenyl)ethyl]-1,3-benzothiazole, Designed Ankyrin Repeat Protein (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wranik, M, Weinert, T, Standfuss, J, Steinmetz, M. | | Deposit date: | 2022-02-21 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | In Vivo Photocontrol of Microtubule Dynamics and Integrity, Migration and Mitosis, by the Potent GFP-Imaging-Compatible Photoswitchable Reagents SBTubA4P and SBTub2M.
J.Am.Chem.Soc., 144, 2022
|
|
7ZEV
 
 | | Free form of extended Cyp33-RRM | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7ZEW
 
 | | Complex Cyp33-RRM : AAUAAA RNA | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E, RNA (5'-R(*AP*AP*UP*AP*AP*A)-3') | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7ZEZ
 
 | | Trimolecular complex Cyp33-RRMdelta alpha : MLL1-PHD3 : H3K4me3 | | Descriptor: | Histone H3, Isoform 3 of Peptidyl-prolyl cis-trans isomerase E, MLL cleavage product N320, ... | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-05-03 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7ZEX
 
 | | Complex Cyp33-RRMdelta alpha : UAAUGUCG RNA | | Descriptor: | Isoform 3 of Peptidyl-prolyl cis-trans isomerase E, RNA (5'-R(*UP*AP*AP*UP*GP*UP*CP*G)-3') | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7YYW
 
 | | Molecular snapshots of drug release from tubulin: 10 nanoseconds after photoactivation. | | Descriptor: | Azo-Combretastatin A4 (trans), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Wranik, M, Weinert, T, Standfuss, J. | | Deposit date: | 2022-02-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Watching the release of a photopharmacological drug from tubulin using time-resolved serial crystallography.
Nat Commun, 14, 2023
|
|
7YZ5
 
 | | Molecular snapshots of drug release from tubulin: 100 milliseconds (steady state) | | Descriptor: | CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wranik, M, Weinert, T, Standfuss, J. | | Deposit date: | 2022-02-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Watching the release of a photopharmacological drug from tubulin using time-resolved serial crystallography.
Nat Commun, 14, 2023
|
|
7YYV
 
 | | Molecular snapshots of drug release from tubulin: 1 nanosecond after photoactivation. | | Descriptor: | Azo-Combretastatin A4 (trans), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Wranik, M, Weinert, T, Standfuss, J. | | Deposit date: | 2022-02-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Watching the release of a photopharmacological drug from tubulin using time-resolved serial crystallography.
Nat Commun, 14, 2023
|
|
7YZ2
 
 | | Molecular snapshots of drug release from tubulin: 10 milliseconds after photoactivation. | | Descriptor: | Designed Ankyrin Repeat Protein (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wranik, M, Weinert, T, Standfuss, J. | | Deposit date: | 2022-02-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Watching the release of a photopharmacological drug from tubulin using time-resolved serial crystallography.
Nat Commun, 14, 2023
|
|
7YYZ
 
 | | Molecular snapshots of drug release from tubulin: 10 microseconds after photoactivation. | | Descriptor: | Azo-Combretastatin A4 (trans), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Wranik, M, Weinert, T, Standfuss, J. | | Deposit date: | 2022-02-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Watching the release of a photopharmacological drug from tubulin using time-resolved serial crystallography.
Nat Commun, 14, 2023
|
|
7YZ6
 
 | | Molecular snapshots of drug release from tubulin: Dark (steady state) | | Descriptor: | Azo-Combretastatin A4 (cis), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Wranik, M, Weinert, T, Standfuss, J. | | Deposit date: | 2022-02-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Watching the release of a photopharmacological drug from tubulin using time-resolved serial crystallography.
Nat Commun, 14, 2023
|
|
