8DMY
 
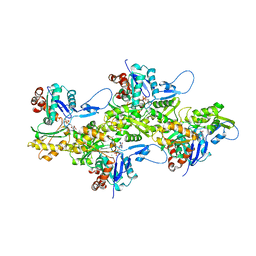 | | Cryo-EM structure of cardiac muscle alpha-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha cardiac muscle 1, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-09 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
9BUJ
 
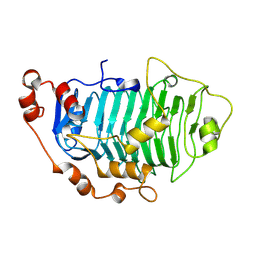 | | Structure of PfPL1 from Pseudoalteromonas fuliginea | | Descriptor: | CALCIUM ION, Pectate lyase | | Authors: | Boraston, A.B, Hobbs, J.K. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The structure of a pectin-active family 1 polysaccharide lyase from the marine bacterium Pseudoalteromonas fuliginea.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8DNF
 
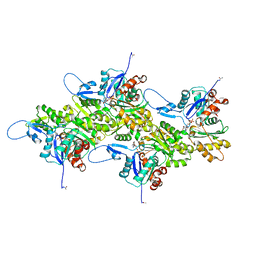 | | Cryo-EM structure of nonmuscle gamma-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 2, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
8DMX
 
 | | Cryo-EM structure of skeletal muscle alpha-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
8DNH
 
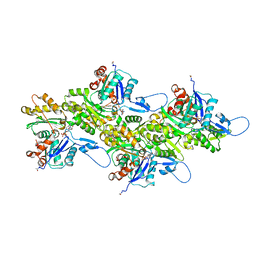 | | Cryo-EM structure of nonmuscle beta-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
7MFL
 
 | |
7MFK
 
 | |
6M8N
 
 | | Endo-fucoidan hydrolase P5AFcnA from glycoside hydrolase family 107 | | Descriptor: | CALCIUM ION, MALONATE ION, P5AFcnA | | Authors: | Boraston, A.B, Vickers, C.J, Abe, K, Salama-Alber, O. | | Deposit date: | 2018-08-22 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
1Y9O
 
 | | 1H NMR Structure of Acylphosphatase from the hyperthermophile Sulfolobus Solfataricus | | Descriptor: | Acylphosphatase | | Authors: | Corazza, A, Rosano, C, Pagano, K, Alverdi, V, Esposito, G, Capanni, C, Bemporad, F, Plakoutsi, G, Stefani, M, Chiti, F, Zuccotti, S, Bolognesi, M, Viglino, P. | | Deposit date: | 2004-12-16 | | Release date: | 2005-11-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, conformational stability, and enzymatic properties of acylphosphatase from the hyperthermophile Sulfolobus solfataricus
Proteins, 62, 2006
|
|
7LK7
 
 | |
7LNP
 
 | |
6L6O
 
 | | Crystal structure of stabilized Rab5a GTPase domain from Leishmania donovani | | Descriptor: | ACETATE ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Arora, A, Zohib, M, Biswal, B.K, Maheshwari, D, Pal, R.K. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the GDP-bound GTPase domain of Rab5a from Leishmania donovani.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8U36
 
 | |
6ALK
 
 | | NMR solution structure of the major beech pollen allergen Fag s 1 | | Descriptor: | Fag s 1 pollen allergen | | Authors: | Moraes, A.H, Asam, A, Almeida, F.C.L, Wallner, M, Ferreira, F, Valente, A.P. | | Deposit date: | 2017-08-08 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for cross-reactivity and conformation fluctuation of the major beech pollen allergen Fag s 1.
Sci Rep, 8, 2018
|
|
8SV0
 
 | |
3LQY
 
 | | Crystal structure of putative isochorismatase hydrolase from Oleispira antarctica | | Descriptor: | GLYCEROL, putative isochorismatase hydrolase | | Authors: | Goral, A, Chruszcz, M, Kagan, O, Cymborowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-10 | | Release date: | 2010-03-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative isochorismatase hydrolase from Oleispira antarctica.
J.Struct.Funct.Genom., 13, 2012
|
|
1W9S
 
 | |
1W9T
 
 | | Structure of a beta-1,3-glucan binding CBM6 from Bacillus halodurans in complex with xylobiose | | Descriptor: | BH0236 PROTEIN, SODIUM ION, alpha-D-xylopyranose, ... | | Authors: | Boraston, A.B, van Bueren, A.L. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Family 6 Carbohydrate Binding Modules Recognize the Non-Reducing End of Beta-1,3-Linked Glucans by Presenting a Unique Ligand Binding Surface
J.Biol.Chem., 280, 2005
|
|
1W9W
 
 | |
1G90
 
 | |
1OH4
 
 | | Structural and thermodynamic dissection of specific mannan recognition by a carbohydrate-binding module | | Descriptor: | BETA-MANNOSIDASE, CALCIUM ION, GLYCEROL, ... | | Authors: | Boraston, A.B, Revett, T.J, Boraston, C.M, Nurizzo, D, Davies, G.J. | | Deposit date: | 2003-05-21 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Thermodynamic Dissection of Specific Mannan Recognition by a Carbohydrate Binding Module, Tmcbm27
Structure, 11, 2003
|
|
1OF3
 
 | | Structural and thermodynamic dissection of specific mannan recognition by a carbohydrate-binding module, TmCBM27 | | Descriptor: | BETA-MANNOSIDASE, CALCIUM ION | | Authors: | Boraston, A.B, Revett, T.J, Boraston, C.M, Nurizzo, D, Davies, G.J. | | Deposit date: | 2003-04-07 | | Release date: | 2003-04-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Thermodynamic Dissection of Specific Mannan Recognition by a Carbohydrate Binding Module, Tmcbm27
Structure, 11, 2003
|
|
1OF4
 
 | | Structural and thermodynamic dissection of specific mannan recognition by a carbohydrate-binding module, TmCBM27 | | Descriptor: | BETA-MANNOSIDASE, CALCIUM ION, GLYCEROL, ... | | Authors: | Boraston, A.B, Revett, T.J, Boraston, C.M, Nurizzo, D, Davies, G.J. | | Deposit date: | 2003-04-07 | | Release date: | 2003-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Thermodynamic Dissection of Specific Mannan Recognition by a Carbohydrate Binding Module, Tmcbm27
Structure, 11, 2003
|
|
7WH4
 
 | |
3K9V
 
 | | Crystal structure of rat mitochondrial P450 24A1 S57D in complex with CHAPS | | Descriptor: | 1,25-dihydroxyvitamin D(3) 24-hydroxylase, mitochondrial, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, ... | | Authors: | Annalora, A.J, Goodin, D.B, Hong, W, Zhang, Q, Johnson, E.F, Stout, C.D. | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CYP24A1, a mitochondrial cytochrome P450 involved in vitamin D metabolism.
J.Mol.Biol., 396, 2010
|
|
