5Y7C
 
 | | Hapalindole A and DMSPP Bound AmbP3 | | Descriptor: | AmbP3, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Hapalindole A | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Y84
 
 | | Hapalindole U and DMSPP Bound AmbP3 | | Descriptor: | AmbP3, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Hapalindole U | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-18 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5YQS
 
 | | Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yaoi, K, Matsuzawa, T, Watanabe, M, Nakamichi, Y. | | Deposit date: | 2017-11-07 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and substrate recognition mechanism of Aspergillus oryzae isoprimeverose-producing enzyme.
J.Struct.Biol., 205, 2019
|
|
5Z43
 
 | | Crystal structure of prenyltransferase AmbP1 apo structure | | Descriptor: | AmbP1, MAGNESIUM ION | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z45
 
 | | Crystal structure of prenyltransferase AmbP1 pH6.5 complexed with GSPP and cis-indolyl vinyl isonitrile | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, AmbP1, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
2ZTU
 
 | | T190A mutant of D-3-hydroxybutyrate dehydrogenase complexed with NAD+ | | Descriptor: | D(-)-3-hydroxybutyrate dehydrogenase, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-09 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
5ZN6
 
 | |
5Z44
 
 | | Crystal structure of prenyltransferase AmbP1 complexed with GSPP | | Descriptor: | AmbP1, GERANYL S-THIOLODIPHOSPHATE, MAGNESIUM ION | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.458 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
2ZTM
 
 | | T190S mutant of D-3-hydroxybutyrate dehydrogenase | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, D(-)-3-hydroxybutyrate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-07 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
3A0A
 
 | | Structure of (PPG)4-OPG-(PPG)4, monoclinic, twinned crystal | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Morimoto, T, Hongo, C, Katagiri, A, Tanaka, Y, Nishino, N. | | Deposit date: | 2009-03-13 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure of (PPG)4-OPG-(PPG)4
To be Published
|
|
2ZZT
 
 | | Crystal structure of the cytosolic domain of the cation diffusion facilitator family protein | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Higuchi, T, Hattori, M, Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2009-02-25 | | Release date: | 2009-08-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.842 Å) | | Cite: | Crystal structure of the cytosolic domain of the cation diffusion facilitator family protein
Proteins, 76, 2009
|
|
5ZN7
 
 | |
5Z46
 
 | | Crystal structure of prenyltransferase AmbP1 pH8 complexed with GSPP and cis-indolyl vinyl isonitrile | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, AmbP1, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
2ZTL
 
 | | Closed conformation of D-3-hydroxybutyrate dehydrogenase complexed with NAD+ and L-3-hydroxybutyrate | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, D(-)-3-hydroxybutyrate dehydrogenase, GLYCEROL, ... | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-07 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
2ZTV
 
 | | The binary complex of D-3-hydroxybutyrate dehydrogenase with NAD+ | | Descriptor: | D(-)-3-hydroxybutyrate dehydrogenase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-09 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
5Y4G
 
 | | Apo Structure of AmbP3 | | Descriptor: | AmbP3 | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-03 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
3AP2
 
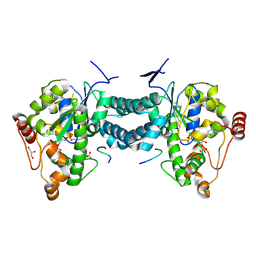 | | Crystal structure of human tyrosylprotein sulfotransferase-2 complexed with PAP,C4 peptide, and phosphate ion | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, C4 peptide, GLYCEROL, ... | | Authors: | Teramoto, T, Fujikawa, Y, Kawaguchi, Y, Kurogi, K, Soejima, M, Adachi, R, Nakanishi, Y, Mishiro-Sato, E, Liu, M.-C, Sakakibara, Y, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2010-10-09 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human tyrosylprotein sulfotransferase-2: Insights into substrate-binding and catalysis of post-translational protein tyrosine sulfation
To be Published
|
|
3AP3
 
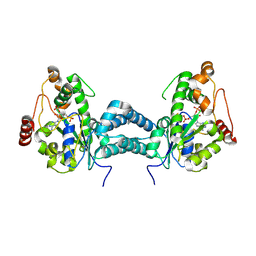 | | Crystal structure of human tyrosylprotein sulfotransferase-2 complexed with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Protein-tyrosine sulfotransferase 2 | | Authors: | Teramoto, T, Fujikawa, Y, Kawaguchi, Y, Kurogi, K, Soejima, M, Adachi, R, Nakanishi, Y, Mishiro-Sato, E, Liu, M.-C, Sakakibara, Y, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2010-10-09 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction.
Nat Commun, 4, 2013
|
|
3AP1
 
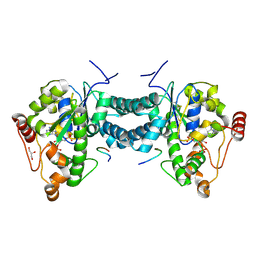 | | Crystal structure of human tyrosylprotein sulfotransferase-2 complexed with PAP and C4 peptide | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, C4 peptide, GLYCEROL, ... | | Authors: | Teramoto, T, Fujikawa, Y, Kawaguchi, Y, Kurogi, K, Soejima, M, Adachi, R, Nakanishi, Y, Mishiro-Sato, E, Liu, M.-C, Sakakibara, Y, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2010-10-09 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction.
Nat Commun, 4, 2013
|
|
2YVX
 
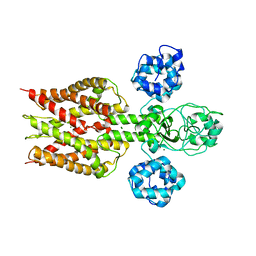 | | Crystal structure of magnesium transporter MgtE | | Descriptor: | MAGNESIUM ION, Mg2+ transporter MgtE | | Authors: | Hattori, M, Tanaka, Y, Fukai, S, Ishitani, R, Nureki, O. | | Deposit date: | 2007-04-18 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the MgtE Mg(2+) transporter
Nature, 448, 2007
|
|
3BIK
 
 | | Crystal Structure of the PD-1/PD-L1 Complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BIS
 
 | | Crystal Structure of the PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7EAP
 
 | | Crystal structure of IpeA-XXXG complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Matsuzawa, T, Watanabe, M, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural basis for the catalytic mechanism of the glycoside hydrolase family 3 isoprimeverose-producing oligoxyloglucan hydrolase from Aspergillus oryzae.
Febs Lett., 596, 2022
|
|
7EDA
 
 | | Structure of monomeric photosystem II | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Yu, H, Hamaguchi, T, Nakajima, Y, Kato, K, kawakami, K, Akita, F, Yonekura, K, Shen, J.R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Cryo-EM structure of monomeric photosystem II at 2.78 angstrom resolution reveals factors important for the formation of dimer.
Biochim Biophys Acta Bioenerg, 1862, 2021
|
|
7D1T
 
 | | Cryo-EM Structure of PSII at 1.95 angstrom resolution | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kato, K, Miyazaki, N, Hamaguchi, T, Nakajima, Y, Akita, F, Yonekura, K, Shen, J.R. | | Deposit date: | 2020-09-15 | | Release date: | 2021-03-31 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (1.95 Å) | | Cite: | High-resolution cryo-EM structure of photosystem II reveals damage from high-dose electron beams.
Commun Biol, 4, 2021
|
|
