4N7W
 
 | | Crystal Structure of the sodium bile acid symporter from Yersinia frederiksenii | | Descriptor: | CITRIC ACID, Transporter, sodium/bile acid symporter family, ... | | Authors: | Zhou, X, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-10-16 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural basis of the alternating-access mechanism in a bile acid transporter.
Nature, 505, 2013
|
|
4MOT
 
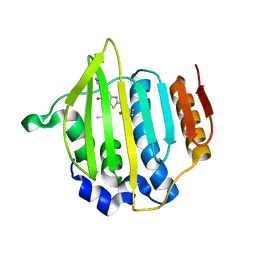 | | Structure of Streptococcus pneumonia pare in complex with AZ13072886 | | Descriptor: | 1-[4-(3-methylbutyl)-5-oxo-6-(pyridin-3-yl)-4,5-dihydro[1,3]thiazolo[5,4-b]pyridin-2-yl]-3-prop-2-en-1-ylurea, Topoisomerase IV subunit B | | Authors: | Ogg, D, Boriack-Sjodin, P.A. | | Deposit date: | 2013-09-12 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thiazolopyridone ureas as DNA gyrase B inhibitors: Optimization of antitubercular activity and efficacy.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N7X
 
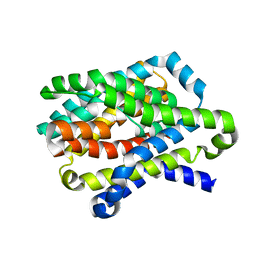 | |
2LNA
 
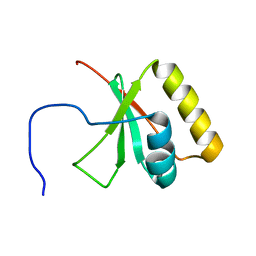 | | Solution NMR Structure of the mitochondrial inner membrane domain (residues 164-251), FtsH_ext, from the paraplegin-like protein AFG3L2 from Homo sapiens, Northeast Structural Genomics Consortium Target HR6741A | | Descriptor: | AFG3-like protein 2 | | Authors: | Ramelot, T.A, Yang, Y, Lee, H, Janua, H, Kohan, E, Shastry, R, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and MD simulations of the AAA protease intermembrane space domain indicates peripheral membrane localization within the hexaoligomer.
Febs Lett., 587, 2013
|
|
8C3W
 
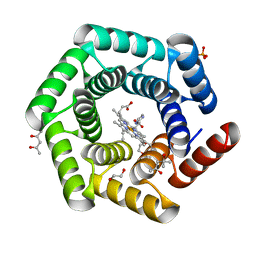 | | Crystal structure of a computationally designed heme binding protein, dnHEM1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ortmayer, M, Levy, C. | | Deposit date: | 2022-12-29 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Heme Enzymes with a Tunable Substrate Binding Pocket Adjacent to an Open Metal Coordination Site.
J.Am.Chem.Soc., 145, 2023
|
|
6VNS
 
 | | Crystal structure of TYK2 kinase with compound 13 | | Descriptor: | (1R,2R)-2-cyano-N-[(1S,5R)-3-(5-fluoro-2-{[1-(2-hydroxyethyl)-1H-pyrazol-4-yl]amino}pyrimidin-4-yl)-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropane-1-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
6VNY
 
 | | Crystal structure of TYK2 kinase with compound 10 | | Descriptor: | N-[(1S,5R)-3-(5-fluoro-2-{[1-(2-hydroxyethyl)-1H-pyrazol-4-yl]amino}pyrimidin-4-yl)-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropanecarboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
8FUI
 
 | | HIV-1 wild type protease with GRL-02519A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
8FUJ
 
 | | HIV-1 wild type protease with GRL-03419A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group and 3,5-difluorophenylmethyl as the P1 group | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
6W8L
 
 | | Crystal structure of JAK1 kinase with compound 10 | | Descriptor: | N-[(1S,5R)-3-(5-fluoro-2-{[1-(2-hydroxyethyl)-1H-pyrazol-4-yl]amino}pyrimidin-4-yl)-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropanecarboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
8DI1
 
 | |
8DI0
 
 | |
6VNX
 
 | | Crystal structure of TYK2 kinase with compound 19 | | Descriptor: | (1S)-2,2-difluoro-N-[(1S,5R,6R)-3-{5-fluoro-2-[(1-methyl-1H-pyrazol-4-yl)amino]pyrimidin-4-yl}-6-methyl-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropane-1-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
8CT5
 
 | |
8CTA
 
 | | Minimal 2:2 Ternary Complex between BI-224436 bound HIV-1 Integrase Catalytic Core Domain Dimer and Carboxy Terminal Domains | | Descriptor: | (2S)-tert-butoxy[4-(2,3-dihydropyrano[4,3,2-de]quinolin-7-yl)-2-methylquinolin-3-yl]acetic acid, 1,2-ETHANEDIOL, Integrase | | Authors: | Gupta, K, Van Duyne, G.D, Eilers, G, Bushman, F.D. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structure of a HIV-1 IN-Allosteric inhibitor complex at 2.93 angstrom resolution: Routes to inhibitor optimization.
Plos Pathog., 19, 2023
|
|
8CT7
 
 | | Catalytic Core Domain of HIV-1 Integrase (F185K) bound with BI-224436 | | Descriptor: | (2S)-tert-butoxy[4-(2,3-dihydropyrano[4,3,2-de]quinolin-7-yl)-2-methylquinolin-3-yl]acetic acid, 1,2-ETHANEDIOL, Integrase, ... | | Authors: | Gupta, K, Van Duyne, G.D, Eilers, G, Bushman, F.D. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of a HIV-1 IN-Allosteric inhibitor complex at 2.93 angstrom resolution: Routes to inhibitor optimization.
Plos Pathog., 19, 2023
|
|
8Q4E
 
 | |
8QK8
 
 | |
2KKQ
 
 | | Solution NMR Structure of the Ig-like C2-type 2 Domain of Human Myotilin. Northeast Structural Genomics Target HR3158. | | Descriptor: | Myotilin | | Authors: | Rossi, P, Shastry, R, Ciccosanti, C, Hamilton, K, Xiao, R, Acton, T.B, Swapna, G.V.T, Nair, R, Everett, J.K, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Ig-like C2-type 2 Domain of Human Myotilin. Northeast Structural Genomics Target HR3158.
To be Published
|
|
5KM4
 
 | |
5KLZ
 
 | |
5KM5
 
 | | Human Histidine Triad Nucleotide Binding Protein 2 (hHint2) triciribine 5'-monoposphate catalytic product complex | | Descriptor: | 5-methyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-1,5-dihydro-1,4,5,6,8-pentaazaacenaphthylen-3-amine, CHLORIDE ION, Histidine triad nucleotide-binding protein 2, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
6VNV
 
 | | Crystal structure of TYK2 kinase with compound 14 | | Descriptor: | (1S,2S)-2-cyano-N-[(1S,5R)-3-(5-fluoro-2-{[1-(2-hydroxyethyl)-1H-pyrazol-4-yl]amino}pyrimidin-4-yl)-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropane-1-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
5KM9
 
 | |
5KM3
 
 | |
