7E6N
 
 | | Crystal structure of HCoV-NL63 3C-like protease,pH5.2 | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8413 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24916625 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2808516 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7F41
 
 | | PARP15 catalytic domain in complex with 3-AMINOBENZAMIDE | | Descriptor: | 3-aminobenzamide, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-06-17 | | Release date: | 2022-06-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.39721382 Å) | | Cite: | Crystal structures of the catalytic domain of human PARP15 in complex with small molecule inhibitors
Biochem.Biophys.Res.Commun., 622, 2022
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
5EGO
 
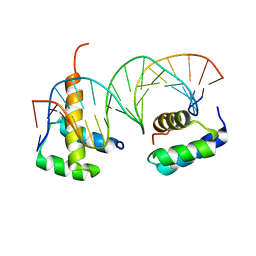 | | HOXB13-MEIS1 heterodimer bound to methylated DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*TP*GP*TP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*AP*CP*AP*GP*TP*TP*TP*TP*AP*(5CM)P*GP*AP*GP*G)-3'), Homeobox protein Hox-B13, ... | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
5EF6
 
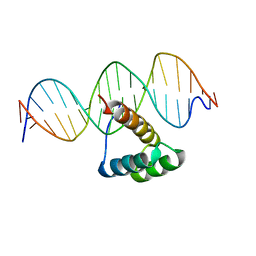 | | Structure of HOXB13 complex with methylated DNA | | Descriptor: | DNA (5'-D(P*GP*GP*AP*CP*CP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*(5CM)P*GP*AP*GP*GP*TP*CP*C)-3'), Homeobox protein Hox-B13 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-23 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
8RJC
 
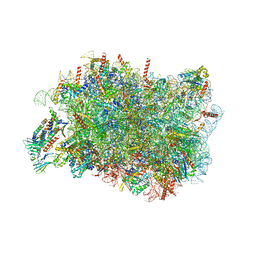 | |
8RJB
 
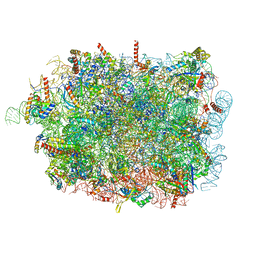 | |
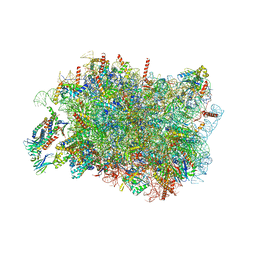
8RJD
 
 | |
7XC3
 
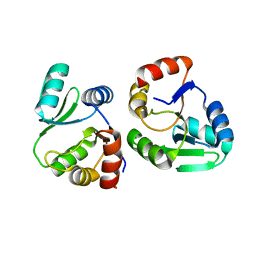 | |
7XC4
 
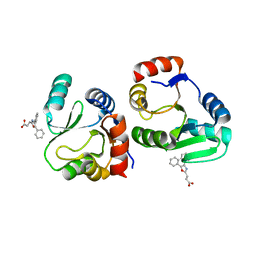 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain 3 (SARS-unique domain-M) in complex with Oxaprozin | | Descriptor: | 3-(4,5-diphenyl-1,3-oxazol-2-yl)propanoic acid, Papain-like protease nsp3 | | Authors: | Li, J, Liu, Y, Gao, J, Ruan, K. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Binding Sites of SARS-CoV-2 Macrodomain 3 Probed by Oxaprozin and Meclomen.
J.Med.Chem., 65, 2022
|
|
8HCK
 
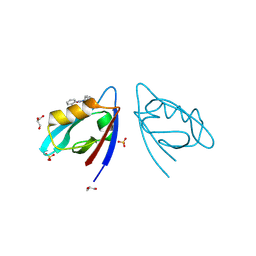 | | NMR fragment-based screening against the two PDZ do-mains of MDA-9 | | Descriptor: | 4-BUTYL-1,2-DIPHENYL-PYRAZOLIDINE-3,5-DIONE, GLYCEROL, SULFATE ION, ... | | Authors: | Tang, H. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitors against Two PDZ Domains of MDA-9 Suppressed Migration of Breast Cancer Cells.
Int J Mol Sci, 24, 2023
|
|
7CRW
 
 | | Cryo-EM structure of rNLRP1-rDPP9 complex | | Descriptor: | Dipeptidyl peptidase 9, NLR family protein 1 | | Authors: | Huang, M.H, Zhang, X.X, Wang, J, Chai, J.J. | | Deposit date: | 2020-08-14 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural and biochemical mechanisms of NLRP1 inhibition by DPP9.
Nature, 592, 2021
|
|
7CRV
 
 | |
7WQJ
 
 | | Crystal structure of MERS main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
4QAO
 
 | | Lysine-ligated cytochrome c with F82H | | Descriptor: | Cytochrome c iso-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Amacher, J.F, Zhu, M.Q, Zhong, F, Pletneva, E.K, Madden, D.R. | | Deposit date: | 2014-05-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Understanding PDZ Affinity and Selectivity: All Residues Considered
Thesis, 2014
|
|
7F43
 
 | | PARP15 catalytic domain in complex with Niraparib | | Descriptor: | 2-{4-[(3S)-piperidin-3-yl]phenyl}-2H-indazole-7-carboxamide, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Zhou, X.L, Zhou, H, Li, J, Zhang, J. | | Deposit date: | 2021-06-17 | | Release date: | 2022-06-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structures of the catalytic domain of human PARP15 in complex with small molecule inhibitors
Biochem.Biophys.Res.Commun., 622, 2022
|
|
