5MUU
 
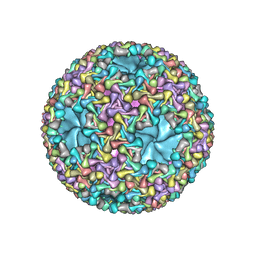 | | dsRNA bacteriophage phi6 nucleocapsid | | Descriptor: | Major inner protein P1, Major outer capsid protein, Packaging enzyme P4 | | Authors: | Sun, Z, El Omari, K, Sun, X, Ilca, S.L, Kotecha, A, Stuart, D.I, Poranen, M.M, Huiskonen, J.T. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Double-stranded RNA virus outer shell assembly by bona fide domain-swapping.
Nat Commun, 8, 2017
|
|
5MUV
 
 | | Atomic structure fitted into a localized reconstruction of bacteriophage phi6 packaging hexamer P4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Packaging enzyme P4 | | Authors: | Sun, Z, El Omari, K, Sun, X, Ilca, S, Kotecha, A, Stuart, D.I, Poranen, M.M, Huiskonen, J.T. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Double-stranded RNA virus outer shell assembly by bona fide domain-swapping.
Nat Commun, 8, 2017
|
|
5MUW
 
 | | Atomic structure of P4 packaging enzyme fitted into a localized reconstruction of bacteriophage phi6 vertex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Packaging enzyme P4 | | Authors: | Sun, Z, El Omari, K, Sun, X, Ilca, S, Kotecha, A, Stuart, D.I, Poranen, M.M, Huiskonen, J.T. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Double-stranded RNA virus outer shell assembly by bona fide domain-swapping.
Nat Commun, 8, 2017
|
|
8R19
 
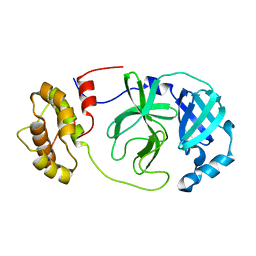 | | SARS-CoV-2 Mpro (Omicron, P132H) free enzyme | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Ibrahim, M, Sun, X, Hilgenfeld, R. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Why is the Omicron main protease of SARS-CoV-2 less stable than its wild-type counterpart? A crystallographic, biophysical, and theoretical study
Hlife, 2, 2024
|
|
8R0V
 
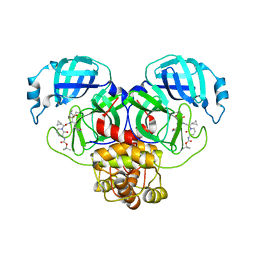 | | SARS-CoV-2 Mpro (Omicron, P132H) in complex with alpha-ketoamide 13b-K at pH 6.5 | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Ibrahim, M, Sun, X, Hilgenfeld, R. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Why is the Omicron main protease of SARS-CoV-2 less stable than its wild-type counterpart? A crystallographic, biophysical, and theoretical study
Hlife, 2, 2024
|
|
8R26
 
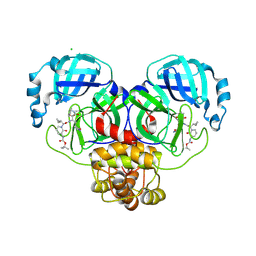 | | SARS-CoV-2 Mpro (Omicron,P132H) in complex with alpha-ketoamide 13b-K at pH 8.5 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Ibrahim, M, Sun, X, Hilgenfeld, R. | | Deposit date: | 2023-11-03 | | Release date: | 2024-11-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Why is the Omicron main protease of SARS-CoV-2 less stable than its wild-type counterpart? A crystallographic, biophysical, and theoretical study
Hlife, 2, 2024
|
|
2RHD
 
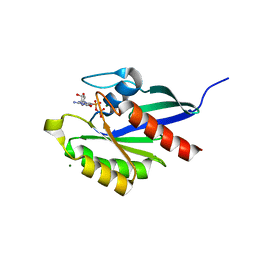 | | Crystal structure of Cryptosporidium parvum small GTPase RAB1A | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Small GTP binding protein rab1a | | Authors: | Dong, A, Xu, X, Lew, J, Lin, Y.H, Khuu, C, Sun, X, Qiu, W, Kozieradzki, I, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Sukumar, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-09 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of Cryptosporidium parvum small GTPase RAB1A.
To be Published
|
|
7F00
 
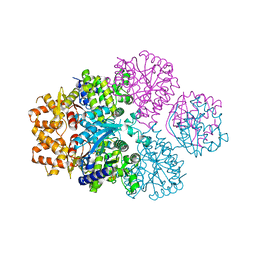 | | Crystal structure of SPD_0310 | | Descriptor: | SULFATE ION, UPF0371 protein SPRM200_0309 | | Authors: | Cao, K, Zhang, T, Li, N, Yang, X, Ding, J, He, Q, Sun, X. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification and Tetramer Structure of Hemin-Binding Protein SPD_0310 Linked to Iron Homeostasis and Virulence of Streptococcus pneumoniae.
Msystems, 7, 2022
|
|
2RCY
 
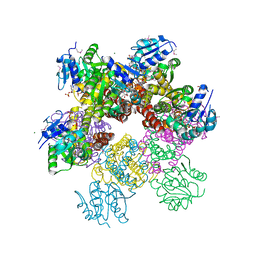 | | Crystal structure of Plasmodium falciparum pyrroline carboxylate reductase (MAL13P1.284) with NADP bound | | Descriptor: | GLYCEROL, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Wernimont, A.K, Lew, J, Lin, Y.H, Ren, H, Sun, X, Khuu, C, Hassanali, A, Wasney, G, Zhao, Y, Kozieradzki, I, Schapira, M, Bochkarev, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Plasmodium falciparum pyrroline carboxylate reductase (MAL13P1.284) with NADP bound.
To be Published
|
|
2R77
 
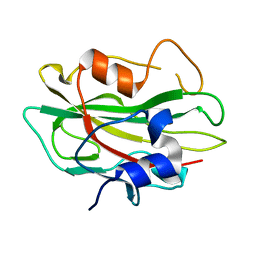 | | Crystal structure of phosphatidylethanolamine-binding protein, pfl0955c, from Plasmodium falciparum | | Descriptor: | Phosphatidylethanolamine-binding protein, putative | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Crombette, L, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-07 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of phosphatidylethanolamine-binding protein, pfl0955c, from Plasmodium falciparum.
To be Published
|
|
5TYT
 
 | | Crystal Structure of the PDZ domain of RhoGEF bound to CXCR2 C-terminal peptide | | Descriptor: | Rho guanine nucleotide exchange factor 11, C-X-C chemokine receptor type 2 chimera | | Authors: | Spellmon, N, Holcomb, J, Niu, A, Choudhary, V, Sun, X, Brunzelle, J, Li, C, Yang, Z. | | Deposit date: | 2016-11-21 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis of PDZ-mediated chemokine receptor CXCR2 scaffolding by guanine nucleotide exchange factor PDZ-RhoGEF.
Biochem. Biophys. Res. Commun., 485, 2017
|
|
5FJ6
 
 | | Structure of the P2 polymerase inside in vitro assembled bacteriophage phi6 polymerase complex | | Descriptor: | MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Ilca, S, Kotecha, A, Sun, X, Poranen, M.P, Stuart, D.I, Huiskonen, J.T. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Localized Reconstruction of Subunits from Electron Cryomicroscopy Images of Macromolecular Complexes.
Nat.Commun., 6, 2015
|
|
3B6N
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase PV003920 from Plasmodium vivax | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ZINC ION | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase PV003920 from Plasmodium vivax.
To be Published
|
|
3BKP
 
 | | Crystal structure of the Toxoplasma gondii cyclophilin, 49.m03261 | | Descriptor: | Cyclophilin, GLYCEROL | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Toxoplasma gondii cyclophilin, 49.m03261.
To be Published
|
|
3BO7
 
 | | Crystal structure of Toxoplasma gondii peptidyl-prolyl cis-trans isomerase, 541.m00136 | | Descriptor: | 1,2-ETHANEDIOL, CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE CYCLOPHILIN-TYPE, ... | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-17 | | Release date: | 2008-02-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Toxoplasma Gondii Peptidyl-Prolyl Cis-Trans Isomerase, 541.M00136.
To be Published
|
|
5FJ7
 
 | | Structure of the P2 polymerase inside in vitro assembled bacteriophage phi6 polymerase complex, with P1 included | | Descriptor: | MAJOR INNER PROTEIN P1, MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Ilca, S, Kotecha, A, Sun, X, Poranen, M.P, Stuart, D.I, Huiskonen, J.T. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Localized Reconstruction of Subunits from Electron Cryomicroscopy Images of Macromolecular Complexes.
Nat.Commun., 6, 2015
|
|
3BE4
 
 | | Crystal structure of Cryptosporidium parvum adenylate kinase cgd5_3360 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, GLYCEROL, ... | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Cryptosporidium parvum adenylate kinase cgd5_3360.
To be Published
|
|
5FJ5
 
 | | Structure of the in vitro assembled bacteriophage phi6 polymerase complex | | Descriptor: | MAJOR INNER PROTEIN P1 | | Authors: | Ilca, S, Kotecha, A, Sun, X, Poranen, M.P, Stuart, D.I, Huiskonen, J.T. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Localized Reconstruction of Subunits from Electron Cryomicroscopy Images of Macromolecular Complexes.
Nat.Commun., 6, 2015
|
|
6LK0
 
 | | Crystal structure of human wild type TRIP13 | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Wang, Y, Huang, J, Li, B, Xue, H, Tricot, G, Hu, L, Xu, Z, Sun, X, Chang, S, Gao, L, Tao, Y, Xu, H, Xie, Y, Xiao, W, Yu, D, Kong, Y, Chen, G, Sun, X, Lian, F, Zhang, N, Wu, X, Mao, Z, Zhan, F, Zhu, W, Shi, J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Small-Molecule Inhibitor Targeting TRIP13 Suppresses Multiple Myeloma Progression.
Cancer Res., 80, 2020
|
|
5IUS
 
 | | Crystal structure of human PD-L1 in complex with high affinity PD-1 mutant | | Descriptor: | CHLORIDE ION, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Pascolutti, R, Sun, X, Kao, J, Maute, R, Ring, A.M, Bowman, G.R, Kruse, A.C. | | Deposit date: | 2016-03-18 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | Structure and Dynamics of PD-L1 and an Ultra-High-Affinity PD-1 Receptor Mutant.
Structure, 24, 2016
|
|
6E67
 
 | | Structure of beta2 adrenergic receptor fused to a Gs peptide | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Beta-2 adrenergic receptor,Endolysin,Guanine nucleotide-binding protein G(s) subunit alpha isoforms short,Beta-2 adrenergic receptor chimera | | Authors: | Liu, X, Xu, X, Hilger, D, Tiemann, J, Liu, H, Du, Y, Hirata, K, Sun, X, Guixa-Gonzalez, R, Mathiesen, J, Hildebrand, P, Kobilka, B. | | Deposit date: | 2018-07-24 | | Release date: | 2019-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Insights into the Process of GPCR-G Protein Complex Formation.
Cell, 177, 2019
|
|
6Y2G
 
 | | Crystal structure (orthorhombic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
6Y2F
 
 | | Crystal structure (monoclinic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
6Y2E
 
 | |
4HBL
 
 | | Crystal structure of AbfR of Staphylococcus epidermidis | | Descriptor: | Transcriptional regulator, MarR family | | Authors: | Liu, X, Sun, X, Gan, J, Lan, L, Yang, C.-G. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Oxidation-sensing Regulator AbfR Regulates Oxidative Stress Responses, Bacterial Aggregation, and Biofilm Formation in Staphylococcus epidermidis.
J.Biol.Chem., 288, 2013
|
|
