5WQ2
 
 | |
6III
 
 | | Crystal structure of an uncharacterized protein | | Descriptor: | ADENOSINE MONOPHOSPHATE, IMIDODIPHOSPHORIC ACID, MAGNESIUM ION, ... | | Authors: | Li, B, Yuan, Z. | | Deposit date: | 2018-10-06 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Crystal structure of an uncharacterized protein
To Be Published
|
|
6INY
 
 | | Crystal structure of an uncharacterized protein | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, UPF0061 protein YdiU | | Authors: | Li, B, Yuan, Z. | | Deposit date: | 2018-10-29 | | Release date: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structure of an uncharacterized protein
To Be Published
|
|
6K20
 
 | | Structure of Apo YdiU | | Descriptor: | MAGNESIUM ION, Protein adenylyltransferase SelO | | Authors: | Li, B, Yang, Y, Ma, Y. | | Deposit date: | 2019-05-13 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of Apo-YdiU
To Be Published
|
|
7D3U
 
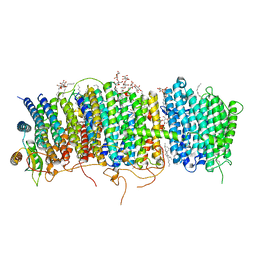 | | Structure of Mrp complex from Dietzia sp. DQ12-45-1b | | Descriptor: | Cation antiporter, DODECYL-BETA-D-MALTOSIDE, Monovalent Na+/H+ antiporter subunit A, ... | | Authors: | Li, B, Zhang, K.D, Wu, X.L, Zhang, X.C. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Dietzia Mrp complex reveals molecular mechanism of this giant bacterial sodium proton pump.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LNA
 
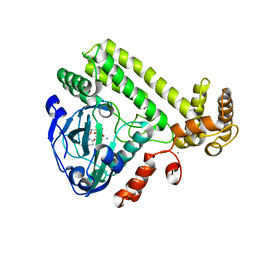 | | YdiU complex with AMPNPP and Mn2+ | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, B, Yang, Y, Ma, Y. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | The YdiU Domain Modulates Bacterial Stress Signaling through Mn 2+ -Dependent UMPylation.
Cell Rep, 32, 2020
|
|
5HKP
 
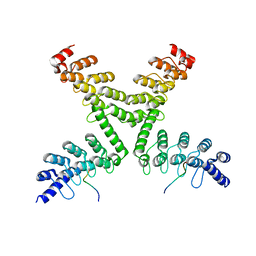 | | Crystal structure of mouse Tankyrase/human TRF1 complex | | Descriptor: | Tankyrase-1, Telomeric repeat-binding factor 1 | | Authors: | Wang, Z, Li, B, Rao, Z, Xu, W. | | Deposit date: | 2016-01-14 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a tankyrase 1-telomere repeat factor 1 complex.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
7TXO
 
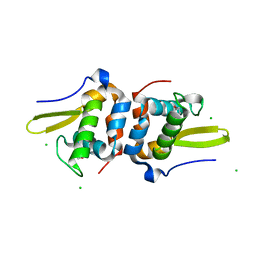 | | Selenomethionine Labeled Structure of RexT | | Descriptor: | CHLORIDE ION, Transcriptional regulator | | Authors: | Bridwell-Rabb, J, Li, B. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and mechanistic basis for redox sensing by the cyanobacterial transcription regulator RexT.
Commun Biol, 5, 2022
|
|
7TXM
 
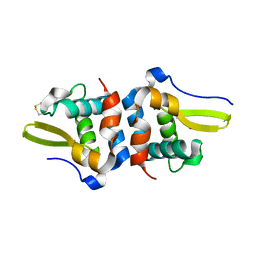 | | Oxidized Structure of RexT | | Descriptor: | HYDROGEN PEROXIDE, Transcriptional regulator | | Authors: | Bridwell-Rabb, J, Li, B. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural and mechanistic basis for redox sensing by the cyanobacterial transcription regulator RexT.
Commun Biol, 5, 2022
|
|
7TXN
 
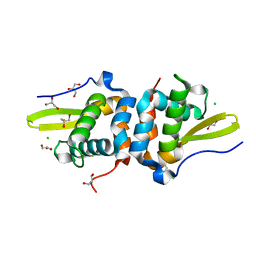 | | Reduced Structure of RexT | | Descriptor: | CHLORIDE ION, GLYCEROL, Transcriptional regulator | | Authors: | Bridwell-Rabb, J, Li, B. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and mechanistic basis for redox sensing by the cyanobacterial transcription regulator RexT.
Commun Biol, 5, 2022
|
|
7M1N
 
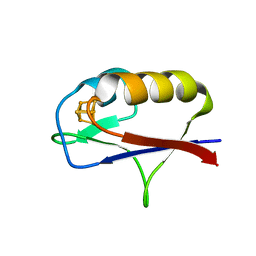 | |
7M1O
 
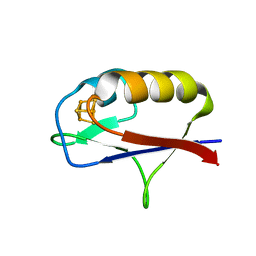 | |
4L7F
 
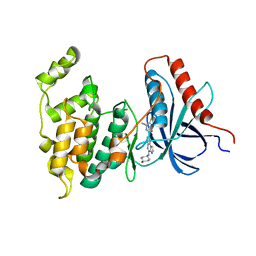 | | Co-crystal Structure of JNK1 and AX13587 | | Descriptor: | Mitogen-activated protein kinase 8, N-[1-(4-fluorophenyl)cyclopropyl]-4-[(trans-4-hydroxycyclohexyl)amino]imidazo[1,2-a]quinoxaline-8-carboxamide | | Authors: | Walter, R.L, Ranieri, G.M, Riggs, A.M, Weissig, H, Li, B, Shreder, K.R. | | Deposit date: | 2013-06-13 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Hit-to-lead optimization and kinase selectivity of imidazo[1,2-a]quinoxalin-4-amine derived JNK1 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7XXH
 
 | | Cryo-EM structure of the purinergic receptor P2Y1R in complex with 2MeSADP and G11 | | Descriptor: | 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate), Guanine nucleotide-binding protein G(11) subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tan, Q, Li, B, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-05-30 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into signal transduction of the purinergic receptors P2Y1R and P2Y12R.
Protein Cell, 14, 2023
|
|
7XXI
 
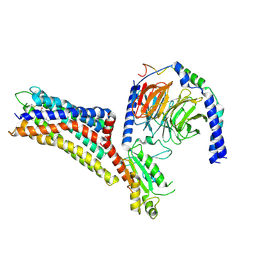 | | Cryo-EM structure of the purinergic receptor P2Y12R in complex with 2MeSADP and Gi | | Descriptor: | 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tan, Q, Li, B, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-05-30 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into signal transduction of the purinergic receptors P2Y1R and P2Y12R.
Protein Cell, 14, 2023
|
|
7DA7
 
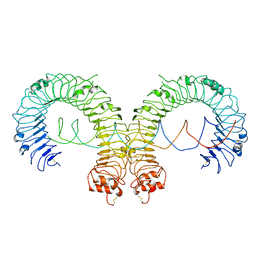 | |
7DAS
 
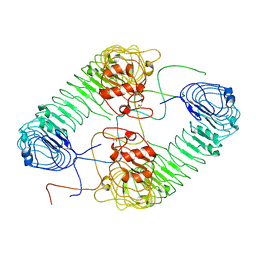 | |
3KUZ
 
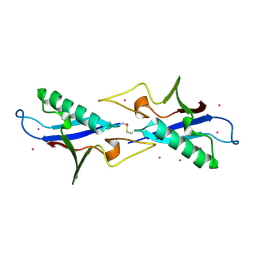 | | Crystal structure of the ubiquitin like domain of PLXNC1 | | Descriptor: | Plexin-C1, UNKNOWN ATOM OR ION | | Authors: | Wang, H, Li, B, Tempel, W, Tong, Y, Guan, X, Zhong, N, Crombet, L, MacKenzie, F, Buck, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ubiquitin like domain of PLXNC1
to be published
|
|
8CJ3
 
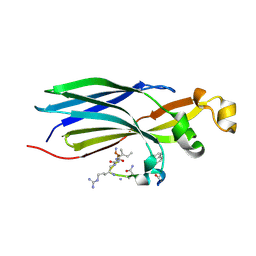 | | Urea-based foldamer inhibitor c3u_7 chimera in complex with ASF1 histone chaperone | | Descriptor: | Histone chaperone ASF1A, c3u_7 chimera inhibitor of histone chaperone ASF1 | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2023-02-11 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
8CJ1
 
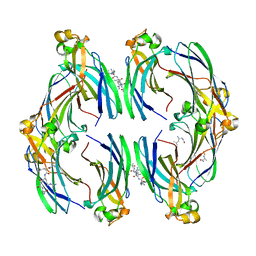 | | Urea-based foldamer inhibitor c3u_3 chimera in complex with ASF1 histone chaperone | | Descriptor: | Histone chaperone ASF1A, c3u_3 chimera inhibitor of histone chaperone ASF1 | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2023-02-11 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.564 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
8CJ2
 
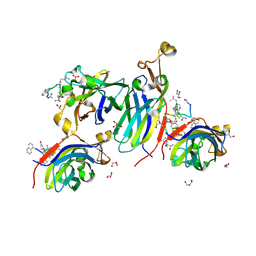 | | Urea-based foldamer inhibitor c3u_5 chimera in complex with ASF1 histone chaperone | | Descriptor: | GLYCEROL, Histone chaperone ASF1A, SULFATE ION, ... | | Authors: | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | Deposit date: | 2023-02-11 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.127 Å) | | Cite: | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
6LK0
 
 | | Crystal structure of human wild type TRIP13 | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Wang, Y, Huang, J, Li, B, Xue, H, Tricot, G, Hu, L, Xu, Z, Sun, X, Chang, S, Gao, L, Tao, Y, Xu, H, Xie, Y, Xiao, W, Yu, D, Kong, Y, Chen, G, Sun, X, Lian, F, Zhang, N, Wu, X, Mao, Z, Zhan, F, Zhu, W, Shi, J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Small-Molecule Inhibitor Targeting TRIP13 Suppresses Multiple Myeloma Progression.
Cancer Res., 80, 2020
|
|
6GCX
 
 | | Focal Adhesion Kinase catalytic domain in complex with irreversible inhibitor | | Descriptor: | 2-[[2-[[4-[[[3,4-bis(oxidanylidene)-2-[2-(propanoylamino)ethylamino]cyclobuten-1-yl]amino]methyl]phenyl]amino]-5-chloranyl-pyrimidin-4-yl]amino]-~{N}-methyl-benzamide, Focal adhesion kinase 1, SULFATE ION | | Authors: | Yen-Pon, E, Li, B, Acebron-Garcia de Eulate, M, Tomkiewicz-Raulet, C, Dawson, J, Lietha, D, Frame, M.C, Coumoul, X, Garbay, C, Etheve-Quelquejeu, M, Chen, H. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of the First Irreversible Inhibitor of Focal Adhesion Kinase.
Acs Chem.Biol., 13, 2018
|
|
6GCR
 
 | | Focal Adhesion Kinase catalytic domain in complex with irreversible inhibitor | | Descriptor: | 2-[[2-[[4-[[[3,4-bis(oxidanylidene)-2-[2-(propanoylamino)ethylamino]cyclobuten-1-yl]amino]methyl]phenyl]amino]-5-chloranyl-pyrimidin-4-yl]amino]-~{N}-methyl-benzamide, Focal adhesion kinase 1 | | Authors: | Yen-Pon, E, Li, B, Acebron-Garcia de Eulate, M, Tomkiewicz-Raulet, C, Dawson, J, Lietha, D, Frame, M.C, Coumoul, X, Garbay, C, Etheve-Quelquejeu, M, Chen, H. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-01 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of the First Irreversible Inhibitor of Focal Adhesion Kinase.
Acs Chem.Biol., 13, 2018
|
|
6GCW
 
 | | Focal Adhesion Kinase catalytic domain in complex with irreversible inhibitor | | Descriptor: | 2-[[5-chloranyl-2-[[4-[[[1-[2-(propanoylamino)ethyl]-1,2,3-triazol-4-yl]methylamino]methyl]phenyl]amino]pyrimidin-4-yl]amino]-~{N}-methyl-benzamide, Focal adhesion kinase 1, SULFATE ION | | Authors: | Yen-Pon, E, Li, B, Acebron-Garcia-de-Eulate, M, Tomkiewicz-Raulet, C, Dawson, J, Lietha, D, Frame, M.C, Coumoul, X, Garbay, C, Etheve-Quelquejeu, M, Chen, H. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of the First Irreversible Inhibitor of Focal Adhesion Kinase.
Acs Chem.Biol., 13, 2018
|
|
