1JU8
 
 | | Solution structure of Leginsulin, a plant hormon | | Descriptor: | Leginsulin | | Authors: | Yamazaki, T, Takaoka, M, Katoh, E, Hanada, K, Sakita, M, Sakata, K, Nishiuchi, Y, Hirano, H. | | Deposit date: | 2001-08-23 | | Release date: | 2003-06-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A possible physiological function and the tertiary structure of a 4-kDa peptide in legumes
EUR.J.BIOCHEM., 270, 2003
|
|
8J12
 
 | | Cryo-EM structure of the AsCas12f-sgRNA-target DNA ternary complex | | Descriptor: | DNA (38-MER), MAGNESIUM ION, RNA (247-MER), ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-12 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.
Cell, 186, 2023
|
|
8J3R
 
 | | Cryo-EM structure of the AsCas12f-HKRA-sgRNAS3-5v7-target DNA | | Descriptor: | DNA (37-MER), DNA (38-MER), MAGNESIUM ION, ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-18 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.
Cell, 186, 2023
|
|
8J1J
 
 | | Cryo-EM structure of the AsCas12f-YHAM-sgRNAS3-5v7-target DNA | | Descriptor: | DNA (38-MER), MAGNESIUM ION, RNA (118-MER), ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-13 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.
Cell, 186, 2023
|
|
5B43
 
 | | Crystal structure of Acidaminococcus sp. Cpf1 in complex with crRNA and target DNA | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cpf1, DNA (34-MER), ... | | Authors: | Yamano, T, Nishimasu, H, Hirano, H, Nakane, T, Ishitani, R, Nureki, O. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA
Cell, 165, 2016
|
|
8H1J
 
 | | Cryo-EM structure of the TnpB-omegaRNA-target DNA ternary complex | | Descriptor: | Non-target strand, RNA-guided DNA endonuclease TnpB, Target strand, ... | | Authors: | Nakagawa, R, Hirano, H, Omura, S, Nureki, O. | | Deposit date: | 2022-10-03 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the transposon-associated TnpB enzyme.
Nature, 616, 2023
|
|
7WSV
 
 | | Cryo-EM structure of the N-terminal deletion mutant of human pannexin-1 in a nanodisc | | Descriptor: | Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2022-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7C7L
 
 | | Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex | | Descriptor: | CRISPR-associated protein Cas14a.1, DNA (40-mer), ZINC ION, ... | | Authors: | Takeda, N.S, Nakagawa, R, Okazaki, S, Hirano, H, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2020-05-26 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the miniature type V-F CRISPR-Cas effector enzyme.
Mol.Cell, 81, 2021
|
|
4ZJ7
 
 | |
4ZDU
 
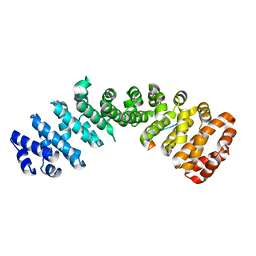 | |
8HIO
 
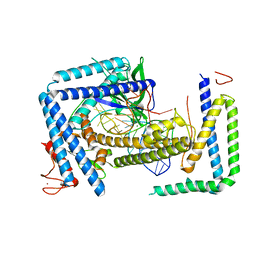 | | Cryo-EM structure of the Cas12m2-crRNA binary complex | | Descriptor: | Cas12m2, MAGNESIUM ION, RNA (56-MER), ... | | Authors: | Omura, N.S, Nakagawa, R, Wu, Y.W, Sudfeld, C, Warren, V.R, Hirano, H, Kusakizako, T, Kise, Y, Lebbink, H.G.J, Itoh, Y, Oost, V.D.J, Nureki, O. | | Deposit date: | 2022-11-21 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8HHM
 
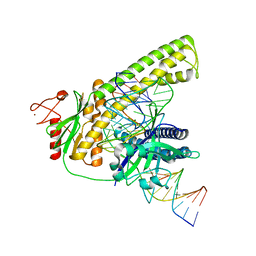 | | Cryo-EM structure of the Cas12m2-crRNA-target DNA ternary complex intermediate state | | Descriptor: | Cas12m2, DNA (36-MER), MAGNESIUM ION, ... | | Authors: | Omura, N.S, Nakagawa, R, Wu, Y.W, Sudfeld, C, Warren, V.R, Hirano, H, Kusakizako, T, Kise, Y, Lebbink, H.G.J, Itoh, Y, Oost, V.D.J, Nureki, O. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8HHL
 
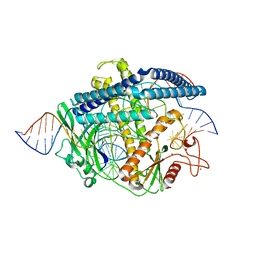 | | Cryo-EM structure of the Cas12m2-crRNA-target DNA full R-loop complex | | Descriptor: | Cas12m2, MAGNESIUM ION, NTS (36-MER), ... | | Authors: | Omura, N.S, Nakagawa, R, Wu, Y.W, Sudfeld, C, Warren, V.R, Hirano, H, Kusakizako, T, Kise, Y, Lebbink, H.G.J, Itoh, Y, Oost, V.D.J, Nureki, O. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7F8O
 
 | | Cryo-EM structure of the C-terminal deletion mutant of human PANX1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8J
 
 | | Cryo-EM structure of human pannexin-1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8N
 
 | | Human pannexin-1 showing a conformational change in the N-terminal domain and blocked pore | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
3AUP
 
 | | Crystal structure of Basic 7S globulin from soybean | | Descriptor: | Basic 7S globulin | | Authors: | Yoshizawa, T, Shimizu, T, Taichi, M, Nishiuchi, Y, Yamabe, M, Shichijo, N, Unzai, S, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-02-14 | | Release date: | 2011-04-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of basic 7S globulin, a xyloglucan-specific endo-beta-1,4-glucanase inhibitor protein-like protein from soybean lacking inhibitory activity against endo-beta-glucanase
Febs J., 278, 2011
|
|
3VL8
 
 | | Crystal structure of XEG | | Descriptor: | SULFATE ION, Xyloglucan-specific endo-beta-1,4-glucanase A | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
3VL9
 
 | | Crystal structure of xeg-xyloglucan | | Descriptor: | Xyloglucan-specific endo-beta-1,4-glucanase A, beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
3VLA
 
 | | Crystal structure of edgp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EDGP | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
3VLB
 
 | | Crystal structure of xeg-edgp | | Descriptor: | EDGP, Xyloglucan-specific endo-beta-1,4-glucanase A | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
5H2V
 
 | |
6AI6
 
 | | Crystal structure of SpCas9-NG | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1, DNA (28-MER), ... | | Authors: | Nishimasu, H, Hirano, S, Ishitani, R, Nureki, O. | | Deposit date: | 2018-08-21 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Engineered CRISPR-Cas9 nuclease with expanded targeting space
Science, 361, 2018
|
|
5JHF
 
 | |
4P1W
 
 | |
