7TGQ
 
 | |
7UAP
 
 | |
7UAQ
 
 | | Structure of the SARS-CoV-2 NTD in complex with C1520, local refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C1520 Fab Heavy Chain, ... | | Authors: | Barnes, C.O. | | Deposit date: | 2022-03-13 | | Release date: | 2022-04-27 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Analysis of memory B cells identifies conserved neutralizing epitopes on the N-terminal domain of variant SARS-Cov-2 spike proteins.
Immunity, 55, 2022
|
|
7UAR
 
 | |
7RKS
 
 | | Structure of the SARS-CoV receptor binding domain in complex with the human neutralizing antibody Fab fragment, C118 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, C118 Antibody Fab Heavy Chain, ... | | Authors: | Jette, C.A, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
7RKU
 
 | | Structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment, C022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C022 Antibody Fab Heavy Chain, C022 Antibody Fab Light Chain, ... | | Authors: | Jette, C.A, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
7RKV
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C118 (State 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C118 Fab Heavy Chain, C118 Fab Light Chain, ... | | Authors: | Barnes, C.O, Jette, C.A, Bjorkman, P.J. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
8FHY
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with neutralizing antibody WRAIR-5021 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MALONATE ION, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2022-12-15 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Diverse array of neutralizing antibodies elicited upon Spike Ferritin Nanoparticle vaccination in rhesus macaques.
Nat Commun, 15, 2024
|
|
8FI9
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody WRAIR-5001 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2022-12-15 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Diverse array of neutralizing antibodies elicited upon Spike Ferritin Nanoparticle vaccination in rhesus macaques.
Nat Commun, 15, 2024
|
|
6UEI
 
 | | Crystal structure of human zinc finger antiviral protein | | Descriptor: | ZINC ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Meagher, J.L, Smith, J.L. | | Deposit date: | 2019-09-21 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of the zinc-finger antiviral protein in complex with RNA reveals a mechanism for selective targeting of CG-rich viral sequences.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6VY2
 
 | |
6VU2
 
 | | M1214_N1 Fab structure | | Descriptor: | M1214 N1 Fab heavy chain, M1214 N1 Fab light chain | | Authors: | Pan, R, Kong, X. | | Deposit date: | 2020-02-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | VSV-Displayed HIV-1 Envelope Identifies Broadly Neutralizing Antibodies Class-Switched to IgG and IgA.
Cell Host Microbe, 27, 2020
|
|
7KZH
 
 | |
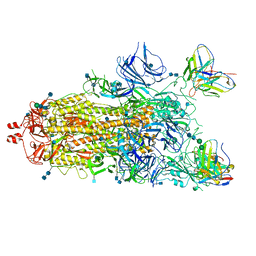
7M6I
 
 | |
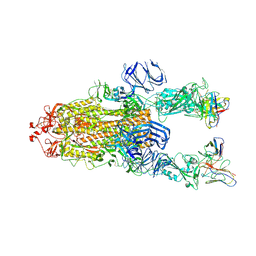
7M6F
 
 | |
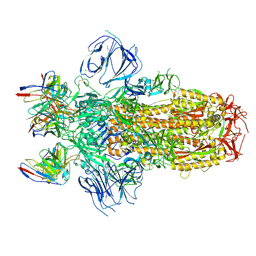
7M6E
 
 | |
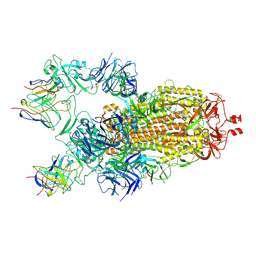
7M6H
 
 | |
7M6G
 
 | |
7M6D
 
 | |
7MRL
 
 | |
