1G3P
 
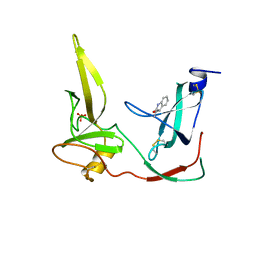 | | CRYSTAL STRUCTURE OF THE N-TERMINAL DOMAINS OF BACTERIOPHAGE MINOR COAT PROTEIN G3P | | Descriptor: | MINOR COAT PROTEIN, SULFATE ION | | Authors: | Lubkowski, J, Hennecke, F, Pluckthun, A, Wlodawer, A. | | Deposit date: | 1997-12-22 | | Release date: | 1998-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The structural basis of phage display elucidated by the crystal structure of the N-terminal domains of g3p.
Nat.Struct.Biol., 5, 1998
|
|
1EML
 
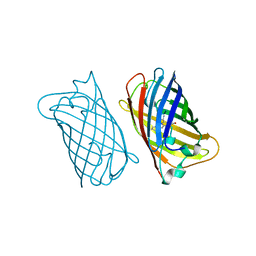 | |
1EMC
 
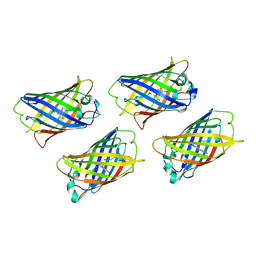 | |
1EMK
 
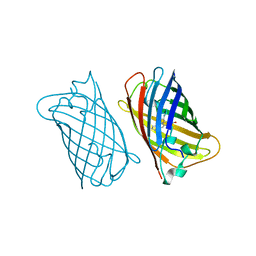 | |
1EMF
 
 | |
1EME
 
 | |
1JH7
 
 | | Semi-reduced Inhibitor-bound Cyclic Nucleotide Phosphodiesterase from Arabidopsis thaliana | | Descriptor: | SULFATE ION, URIDINE-2',3'-VANADATE, cyclic phosphodiesterase | | Authors: | Hofmann, A, Grella, M, Botos, I, Filipowicz, W, Wlodawer, A. | | Deposit date: | 2001-06-27 | | Release date: | 2002-02-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the semireduced and inhibitor-bound forms of cyclic nucleotide phosphodiesterase from Arabidopsis thaliana.
J.Biol.Chem., 277, 2002
|
|
1EMM
 
 | |
1JH6
 
 | | Semi-reduced Cyclic Nucleotide Phosphodiesterase from Arabidopsis thaliana | | Descriptor: | SULFATE ION, cyclic phosphodiesterase | | Authors: | Hofmann, A, Grella, M, Botos, I, Filipowicz, W, Wlodawer, A. | | Deposit date: | 2001-06-27 | | Release date: | 2002-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the semireduced and inhibitor-bound forms of cyclic nucleotide phosphodiesterase from Arabidopsis thaliana.
J.Biol.Chem., 277, 2002
|
|
1I8J
 
 | | CRYSTAL STRUCTURE OF PORPHOBILINOGEN SYNTHASE COMPLEXED WITH THE INHIBITOR 4,7-DIOXOSEBACIC ACID | | Descriptor: | 4,7-DIOXOSEBACIC ACID, MAGNESIUM ION, PORPHOBILINOGEN SYNTHASE, ... | | Authors: | Kervinen, J, Jaffe, E.K, Stauffer, F, Neier, R, Wlodawer, A, Zdanov, A. | | Deposit date: | 2001-03-14 | | Release date: | 2001-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic basis for suicide inactivation of porphobilinogen synthase by 4,7-dioxosebacic acid, an inhibitor that shows dramatic species selectivity.
Biochemistry, 40, 2001
|
|
1BET
 
 | | NEW PROTEIN FOLD REVEALED BY A 2.3 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF NERVE GROWTH FACTOR | | Descriptor: | BETA-NERVE GROWTH FACTOR | | Authors: | Mcdonald, N.Q, Lapatto, R, Murray-Rust, J, Gunning, J, Wlodawer, A, Blundell, T.L. | | Deposit date: | 1993-04-08 | | Release date: | 1994-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New protein fold revealed by a 2.3-A resolution crystal structure of nerve growth factor.
Nature, 354, 1991
|
|
1CXQ
 
 | | ATOMIC RESOLUTION ASV INTEGRASE CORE DOMAIN FROM AMMONIUM SULFATE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AVIAN SARCOMA VIRUS INTEGRASE, GLYCEROL | | Authors: | Lubkowski, J, Dauter, Z, Yang, F, Alexandratos, J, Merkel, G, Skalka, A.M, Wlodawer, A. | | Deposit date: | 1999-08-30 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Atomic resolution structures of the core domain of avian sarcoma virus integrase and its D64N mutant.
Biochemistry, 38, 1999
|
|
1CZB
 
 | | ATOMIC RESOLUTION ASV INTEGRASE CORE DOMAIN FROM HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AVIAN SARCOMA VIRUS INTEGRASE | | Authors: | Lubkowski, J, Dauter, Z, Yang, F, Alexandratos, J, Merkel, G, Skalka, A.M, Wlodawer, A. | | Deposit date: | 1999-09-01 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Atomic resolution structures of the core domain of avian sarcoma virus integrase and its D64N mutant.
Biochemistry, 38, 1999
|
|
1CZ9
 
 | | ATOMIC RESOLUTION ASV INTEGRASE CORE DOMAIN (D64N) FROM CITRATE | | Descriptor: | AVIAN SARCOMA VIRUS INTEGRASE, CITRIC ACID, SULFATE ION | | Authors: | Lubkowski, J, Dauter, Z, Yang, F, Alexandratos, J, Merkel, G, Skalka, A.M, Wlodawer, A. | | Deposit date: | 1999-09-01 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic resolution structures of the core domain of avian sarcoma virus integrase and its D64N mutant.
Biochemistry, 38, 1999
|
|
1DOK
 
 | | MONOCYTE CHEMOATTRACTANT PROTEIN 1, P-FORM | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 1, SULFATE ION | | Authors: | Lubkowski, J, Bujacz, G, Boque, L, Wlodawer, A, Domaille, P.J, Handel, T.M. | | Deposit date: | 1996-11-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions.
Nat.Struct.Biol., 4, 1997
|
|
1DOL
 
 | | MONOCYTE CHEMOATTRACTANT PROTEIN 1, I-FORM | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 1 | | Authors: | Lubkowski, J, Bujacz, G, Boque, L, Wlodawer, A. | | Deposit date: | 1996-11-22 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions.
Nat.Struct.Biol., 4, 1997
|
|
1ASW
 
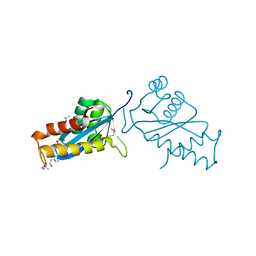 | | AVIAN SARCOMA VIRUS INTEGRASE CATALYTIC CORE DOMAIN CRYSTALLIZED FROM 20% PEG 4000, 10% ISOPROPANOL, HEPES PH 7.5 USING SELENOMETHIONINE SUBSTITUTED PROTEIN; DATA COLLECTED AT-165 DEGREES C | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AVIAN SARCOMA VIRUS INTEGRASE, ISOPROPYL ALCOHOL | | Authors: | Bujacz, G, Jaskolski, M, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1995-08-25 | | Release date: | 1995-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structure of the catalytic domain of avian sarcoma virus integrase.
J.Mol.Biol., 253, 1995
|
|
1ASV
 
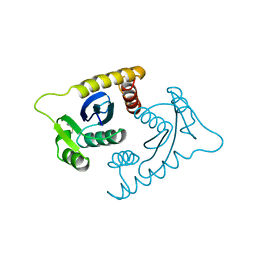 | |
1BG8
 
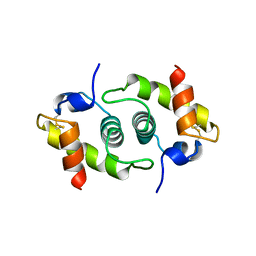 | |
1HIV
 
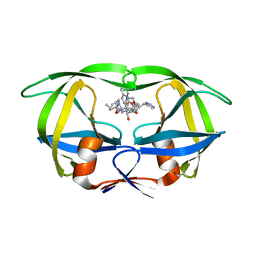 | | CRYSTAL STRUCTURE OF A COMPLEX OF HIV-1 PROTEASE WITH A DIHYDROETHYLENE-CONTAINING INHIBITOR: COMPARISONS WITH MOLECULAR MODELING | | Descriptor: | 4-[(2R)-3-{[(1S,2S,3R,4S)-1-(cyclohexylmethyl)-2,3-dihydroxy-5-methyl-4-({(1S,2R)-2-methyl-1-[(pyridin-2-ylmethyl)carba moyl]butyl}carbamoyl)hexyl]amino}-2-{[(naphthalen-1-yloxy)acetyl]amino}-3-oxopropyl]-1H-imidazol-3-ium, HIV-1 PROTEASE | | Authors: | Thanki, N, Wlodawer, A. | | Deposit date: | 1992-02-12 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a complex of HIV-1 protease with a dihydroxyethylene-containing inhibitor: comparisons with molecular modeling.
Protein Sci., 1, 1992
|
|
1K2A
 
 | | Modified Form of Eosinophil-derived Neurotoxin | | Descriptor: | SULFATE ION, eosinophil-derived neurotoxin | | Authors: | Chang, C, Newton, D.L, Rybak, S.M, Wlodawer, A. | | Deposit date: | 2001-09-26 | | Release date: | 2002-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystallographic and functional studies of a modified form of eosinophil-derived neurotoxin (EDN) with novel biological activities.
J.Mol.Biol., 317, 2002
|
|
1RTA
 
 | |
1RTB
 
 | |
7RY7
 
 | |
7BGU
 
 | | Mason-Pfizer Monkey Virus Protease mutant C7A/D26N/C106A in complex with peptidomimetic inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PENTAETHYLENE GLYCOL, peptidomimetic inhibitor | | Authors: | Wosicki, S, Gilski, M, Kazmierczyk, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2021-01-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.433 Å) | | Cite: | Crystal structures of inhibitor complexes of M-PMV protease with visible flap loops.
Protein Sci., 30, 2021
|
|
