5LDF
 
 | |
5Y27
 
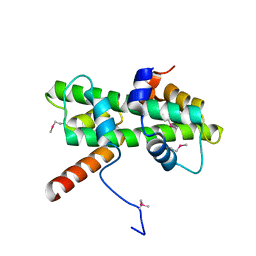 | | Crystal structure of Se-Met Dpb4-Dpb3 | | Descriptor: | DNA polymerase epsilon subunit D, GLYCEROL, Putative transcription factor C16C4.22 | | Authors: | Li, Y, Gao, F, Su, M, Zhang, F.B, Chen, Y.H. | | Deposit date: | 2017-07-24 | | Release date: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coordinated regulation of heterochromatin inheritance by Dpb3-Dpb4 complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7YSF
 
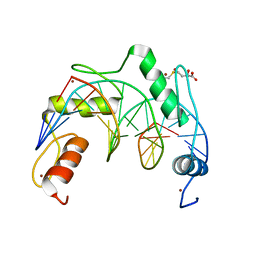 | | Crystal structure of ZNF524 ZF1-4 in complex with telomeric DNA | | Descriptor: | D-MALATE, DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*CP*C)-3'), ... | | Authors: | Li, F.D, Xu, Z.Y. | | Deposit date: | 2022-08-12 | | Release date: | 2023-08-16 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | ZNF524 directly interacts with telomeric DNA and supports telomere integrity.
Nat Commun, 14, 2023
|
|
5Y26
 
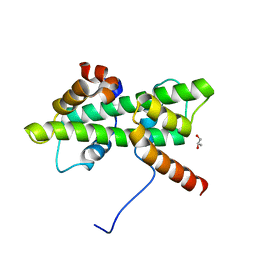 | | Crystal structure of native Dpb4-Dpb3 | | Descriptor: | DNA polymerase epsilon subunit D, GLYCEROL, Putative transcription factor C16C4.22 | | Authors: | Chen, Y.H, Li, Y, Gao, F. | | Deposit date: | 2017-07-24 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Coordinated regulation of heterochromatin inheritance by Dpb3-Dpb4 complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4M14
 
 | |
4M15
 
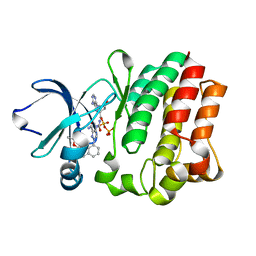 | |
4M12
 
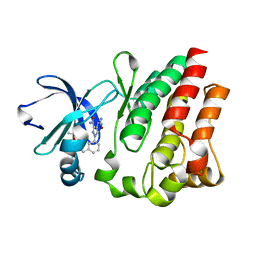 | |
4M0Z
 
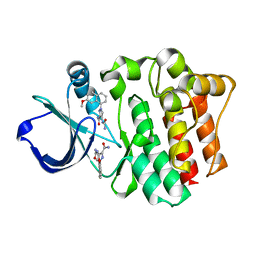 | |
4M0Y
 
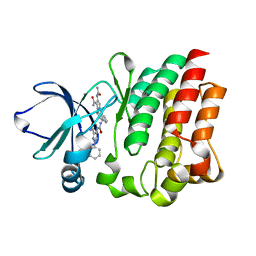 | |
4M13
 
 | |
7U23
 
 | |
6V6R
 
 | | Crystal Structure of a Bromine Derivatized Self-Assembling DNA Crystal Scaffold with Rhombohedral Symmetry. | | Descriptor: | CACODYLATE ION, DNA (5'-D(*CP*AP*CP*TP*GP*AP*CP*TP*CP*AP*TP*GP*CP*TP*CP*AP*TP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*AP*GP*AP*TP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Self-Assembled Rhombohedral DNA Crystal Scaffold with Tunable Cavity Sizes and High-Resolution Structural Detail.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5H7U
 
 | | NMR structure of eIF3 36-163 | | Descriptor: | Eukaryotic translation initiation factor 3 subunit C | | Authors: | Nagata, T, Obayashi, E. | | Deposit date: | 2016-11-21 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular Landscape of the Ribosome Pre-initiation Complex during mRNA Scanning: Structural Role for eIF3c and Its Control by eIF5.
Cell Rep, 18, 2017
|
|
3H0Q
 
 | |
3H0J
 
 | |
7T9U
 
 | | Crystal structure of hSTING with an agonist (SHR169224) | | Descriptor: | (3S,4S)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4-(prop-2-en-1-yl)-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
7T9V
 
 | | Crystal structure of hSTING with the agonist, SHR171032 | | Descriptor: | (3S,4S)-4-(3-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methoxy-1H-benzimidazol-1-yl}propyl)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
7QHE
 
 | | Human Butyrylcholinesterase in complex with (S)-1-(4-((naphthalen-1-yl)carbamoyl)benzyl)-N-(3-((1,2,3,4-tetrahydroacridin-9-yl)amino)propyl)piperidine-3-carboxamide | | Descriptor: | (3~{S})-1-[[4-(naphthalen-1-ylcarbamoyl)phenyl]methyl]-~{N}-[3-(1,2,3,4-tetrahydroacridin-9-ylamino)propyl]piperidine-3-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Jing, L, Zhan, P, Liu, X, Nachon, F. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Rapid discovery and crystallography study of highly potent and selective butylcholinesterase inhibitors based on oxime-containing libraries and conformational restriction strategies.
Bioorg.Chem., 134, 2023
|
|
7QHD
 
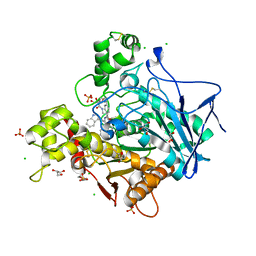 | | Human Butyrylcholinesterase in complex with (S)-1-(4-((2-(1H-indol-3-yl)ethyl)carbamoyl)benzyl)-N-(3-((1,2,3,4-tetrahydroacridin-9-yl)amino)propyl)piperidine-3-carboxamide | | Descriptor: | (3~{S})-1-[[4-[2-(1~{H}-indol-3-yl)ethylcarbamoyl]phenyl]methyl]-~{N}-[3-(1,2,3,4-tetrahydroacridin-9-ylamino)propyl]piperidine-3-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Jing, L, Zhan, P, Liu, X, Nachon, F. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Rapid discovery and crystallography study of highly potent and selective butylcholinesterase inhibitors based on oxime-containing libraries and conformational restriction strategies.
Bioorg.Chem., 134, 2023
|
|
7RN0
 
 | |
7RN1
 
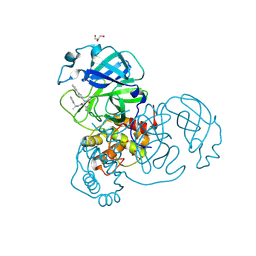 | |
3H0S
 
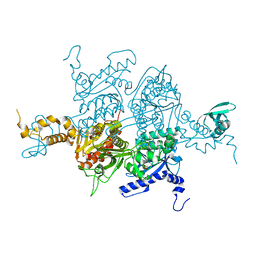 | |
5U5S
 
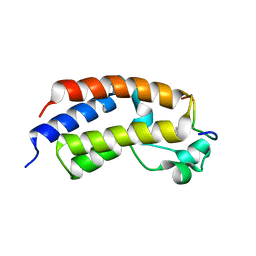 | |
5JYN
 
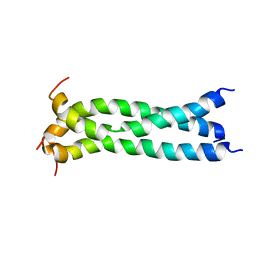 | | Structure of the transmembrane domain of HIV-1 gp41 in bicelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Dev, J, Fu, Q, Park, D, Chen, B, Chou, J.J. | | Deposit date: | 2016-05-14 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for membrane anchoring of HIV-1 envelope spike.
Science, 353, 2016
|
|
7PJR
 
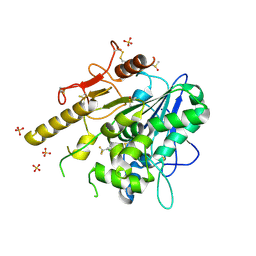 | | Notum_ARUK3000438 | | Descriptor: | 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-1,2,3-triazole, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Vecchia, L, Zhao, Y, Fish, P, Jones, E.Y. | | Deposit date: | 2021-08-24 | | Release date: | 2022-09-07 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Design of a Potent, Selective, and Brain-Penetrant Inhibitor of Wnt-Deactivating Enzyme Notum by Optimization of a Crystallographic Fragment Hit.
J.Med.Chem., 65, 2022
|
|
