8BGG
 
 | | Cryo-EM structure of SARS-CoV-2 spike (Omicron BA.1 variant) in complex with nanobody W25 (map 5, focus refinement on RBD, W25 and adjacent NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody W25, Spike glycoprotein, ... | | Authors: | Modhiran, N, Lauer, S, Spahn, C.M.T, Watterson, D, Schwefel, D. | | Deposit date: | 2022-10-27 | | Release date: | 2023-03-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (6.04 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 spike (Omicron BA.1 variant) in complex with nanobody W25 (map 5, focus refinement on RBD, W25 and adjacent NTD)
To Be Published
|
|
5KAM
 
 | |
5K2E
 
 | | Structure of NNQQNY from yeast prion Sup35 with zinc acetate determined by MicroED | | Descriptor: | ACETIC ACID, Eukaryotic peptide chain release factor GTP-binding subunit, ZINC ION | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4ZA5
 
 | | Structure of A. niger Fdc1 with the prenylated-flavin cofactor in the iminium and ketimine forms. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 1-deoxy-5-O-phosphono-1-[(10aR)-2,2,3,4-tetramethyl-8,10-dioxo-1,2,8,9,10,10a-hexahydro-6H-indeno[1,7-ef]pyrimido[4,5-b][1,4]diazepin-6-yl]-D-ribitol, Fdc1, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
4ZA4
 
 | | Structure of A. niger Fdc1 with the prenylated-flavin cofactor in the iminium form. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Fdc1, MANGANESE (II) ION, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
4ZAF
 
 | |
4ZAY
 
 | | Structure of UbiX E49Q in complex with a covalent adduct between dimethylallyl monophosphate and reduced FMN | | Descriptor: | 1-deoxy-1-[7,8-dimethyl-5-(3-methylbut-2-en-1-yl)-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono -D-ribitol, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
1LGR
 
 | |
6M8H
 
 | | Crystal Structure of the R208Q mutant of G(i) subunit alpha-1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, MAGNESIUM ION | | Authors: | Mascarenhas, R, Goossens, J, Leverson, B, Kothawala, S, Ballicora, M, Olsen, K, de freitas, D, Liu, D. | | Deposit date: | 2018-08-21 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | FUNCTIONAL CONSEQUENCES OF ONCOGENIC MUTATIONS IN THE SWITCH II REGION OF Galphai1 and Galphas PROTEINS
To Be Published
|
|
8OVY
 
 | | Structure of analogue of superfolded GFP | | Descriptor: | Green fluorescent protein | | Authors: | Dunkelmann, D, Fiedler, M, Bellini, D, Alvira, C.P, Chin, J.W. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.537 Å) | | Cite: | Adding alpha , alpha-disubstituted and beta-linked monomers to the genetic code of an organism.
Nature, 625, 2024
|
|
1R0Q
 
 | | Characterization of the conversion of the malformed, recombinant cytochrome rc552 to a 2-formyl-4-vinyl (Spirographis) heme | | Descriptor: | 2-FORMYL-PROTOPORPHRYN IX, Cytochrome c-552 | | Authors: | Fee, J.A, Todaro, T.R, Luna, E, Sanders, D, Hunsicker-Wang, L.M, Patel, K.M, Bren, K.L, Gomez-Moran, E, Hill, M.G, Ai, J, Loehr, T.M, Oertling, W.A, Williams, P.A, Stout, C.D, McRee, D, Pastuszyn, A. | | Deposit date: | 2003-09-22 | | Release date: | 2004-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Cytochrome rC552, formed during expression of the truncated, Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme.
Biochemistry, 43, 2004
|
|
3TRA
 
 | |
4ZIB
 
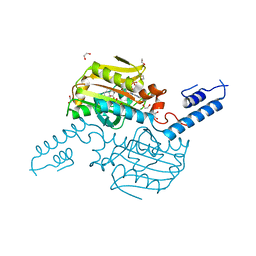 | | Crystal Structure of the C-terminal domain of PylRS mutant bound with 3-benzothienyl-l-alanine and ATP | | Descriptor: | 1,2-ETHANEDIOL, 3-(1-benzothiophen-3-yl)-L-alanine, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Nakamura, A, Guo, L.T, Wang, Y.S, Soll, D. | | Deposit date: | 2015-04-28 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Probing the active site tryptophan of Staphylococcus aureus thioredoxin with an analog.
Nucleic Acids Res., 43, 2015
|
|
4ZAA
 
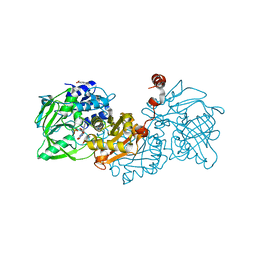 | | Structure of A. niger Fdc1 in complex with 4-vinyl guaiacol | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 1-deoxy-5-O-phosphono-1-[(10aR)-2,2,3,4-tetramethyl-8,10-dioxo-1,2,8,9,10,10a-hexahydro-6H-indeno[1,7-ef]pyrimido[4,5-b][1,4]diazepin-6-yl]-D-ribitol, 2-Methoxy-4-vinylphenol, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
4ETC
 
 | | Lysozyme, room temperature, 24 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
1CVL
 
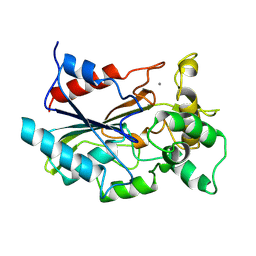 | | CRYSTAL STRUCTURE OF BACTERIAL LIPASE FROM CHROMOBACTERIUM VISCOSUM ATCC 6918 | | Descriptor: | CALCIUM ION, TRIACYLGLYCEROL HYDROLASE | | Authors: | Lang, D.A, Hofmann, B, Haalck, L, Hecht, H.-J, Spener, F, Schmid, R.D, Schomburg, D. | | Deposit date: | 1997-01-09 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a bacterial lipase from Chromobacterium viscosum ATCC 6918 refined at 1.6 angstroms resolution.
J.Mol.Biol., 259, 1996
|
|
1QYZ
 
 | | Characterization of the malformed, recombinant cytochrome rC552 | | Descriptor: | 2-ACETYL-PROTOPORPHYRIN IX, Cytochrome c-552 | | Authors: | Fee, J.A, Todaro, T.R, Luna, E, Sanders, D, Hunsicker-Wang, L.M, Patel, K.M, Bren, K.L, Gomez-Moran, E, Hill, M.G, Ai, J, Loehr, T.M, Oertling, W.A, Williams, P.A, Stout, C.D, McRee, D, Pastuszyn, A. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cytochrome rC552, formed during expression of the truncated, Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme.
Biochemistry, 43, 2004
|
|
1IHP
 
 | | STRUCTURE OF PHOSPHOMONOESTERASE | | Descriptor: | PHYTASE, SULFATE ION | | Authors: | Kostrewa, D. | | Deposit date: | 1997-02-04 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of phytase from Aspergillus ficuum at 2.5 A resolution.
Nat.Struct.Biol., 4, 1997
|
|
8PK4
 
 | | Cryo EM structure of the type 5A polymorph of alpha-synuclein. | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
4ZA9
 
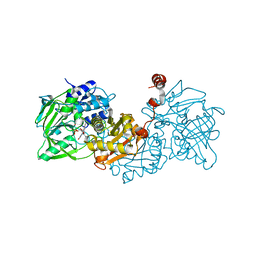 | | Structure of A. niger fdc1 in complex with a phenylpyruvate derived adduct to the prenylated flavin cofactor | | Descriptor: | 1-deoxy-5-O-phosphono-1-[(1S)-3,3,4,5-tetramethyl-9,11-dioxo-1-(phenylacetyl)-2,3,8,9,10,11-hexahydro-1H,7H-quinolino[1 ,8-fg]pteridin-7-yl]-D-ribitol, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
4ZA0
 
 | | Structure of Human Enolase 2 in complex with Phosphonoacetohydroxamate | | Descriptor: | Gamma-enolase, MAGNESIUM ION, PHOSPHONOACETOHYDROXAMIC ACID | | Authors: | Leonard, P.G, Maxwell, D, Czako, B, Muller, F.L. | | Deposit date: | 2015-04-13 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | SF2312 is a natural phosphonate inhibitor of enolase.
Nat.Chem.Biol., 12, 2016
|
|
4ZAD
 
 | | Structure of C. dubliensis Fdc1 with the prenylated-flavin cofactor in the iminium form. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Fdc1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
4ZAV
 
 | | UbiX in complex with a covalent adduct between dimethylallyl monophosphate and reduced FMN | | Descriptor: | 1-deoxy-1-[7,8-dimethyl-5-(3-methylbut-2-en-1-yl)-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono -D-ribitol, PHOSPHATE ION, SODIUM ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
5K2G
 
 | |
5K51
 
 | |
