8JNU
 
 | |
6MRR
 
 | | De novo designed protein Foldit1 | | Descriptor: | Foldit1 | | Authors: | Koepnick, B, Bick, M.J, Estep, R.D, Kleinfelter, S, Wei, L, Baker, D. | | Deposit date: | 2018-10-15 | | Release date: | 2019-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6MRS
 
 | |
7Y1G
 
 | | Crystal structure of human PRKACA complexed with DS01080522 | | Descriptor: | 1-chloranyl-~{N}-[(~{S})-(3-chloranyl-4-cyano-phenyl)-[(2~{R},4~{S})-4-oxidanylpyrrolidin-2-yl]methyl]-7-methoxy-isoquinoline-6-carboxamide, ZINC ION, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Suzuki, M, Ubukata, O, Toyoda, A. | | Deposit date: | 2022-06-08 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel protein kinase cAMP-Activated Catalytic Subunit Alpha (PRKACA) inhibitor shows anti-tumor activity in a fibrolamellar hepatocellular carcinoma model.
Biochem.Biophys.Res.Commun., 621, 2022
|
|
7XZK
 
 | |
7YB3
 
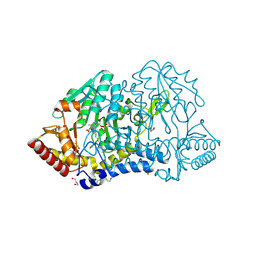 | | SufS with D-cysteine for 1 min | | Descriptor: | Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-06-28 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Snapshots of thiazolidine fomration of PLP-dependent enzyme SufS
to be published
|
|
3WZU
 
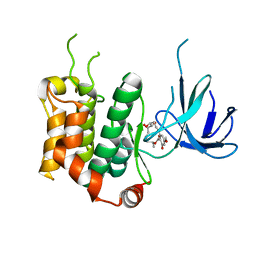 | | THE STRUCTURE OF MAP2K7 IN COMPLEX WITH 5Z-7-oxozeaenol | | Descriptor: | (3S,5Z,8S,9S,11E)-8,9,16-trihydroxy-14-methoxy-3-methyl-3,4,9,10-tetrahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Sogabe, Y, Hashimoto, Y, Matsumoto, T, Kinoshita, T. | | Deposit date: | 2014-10-07 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | 5Z-7-Oxozeaenol covalently binds to MAP2K7 at Cys218 in an unprecedented manner.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5ZC9
 
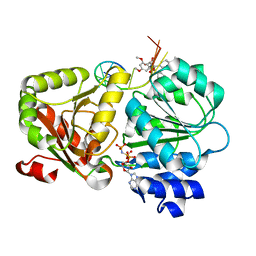 | | Crystal structure of the human eIF4A1-ATP analog-RocA-polypurine RNA complex | | Descriptor: | (1R,2R,3S,3aR,8bS)-6,8-dimethoxy-3a-(4-methoxyphenyl)-N,N-dimethyl-1,8b-bis(oxidanyl)-3-phenyl-2,3-dihydro-1H-cyclopenta[b][1]benzofuran-2-carboxamide, Eukaryotic initiation factor 4A-I, MAGNESIUM ION, ... | | Authors: | Iwasaki, W, Takahashi, M, Sakamoto, A, Iwasaki, S, Ito, T. | | Deposit date: | 2018-02-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Translation Inhibitor Rocaglamide Targets a Bimolecular Cavity between eIF4A and Polypurine RNA.
Mol. Cell, 73, 2019
|
|
6NUK
 
 | | De novo designed protein Ferredog-Diesel | | Descriptor: | Ferredog-Diesel | | Authors: | Koepnick, B, Bick, M.J, DiMaio, F, Norgard-Solano, T, Baker, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
3VRY
 
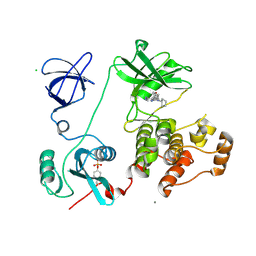 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 4-Amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl-cyclopentane | | Descriptor: | 4-Amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl-cyclopentane, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS7
 
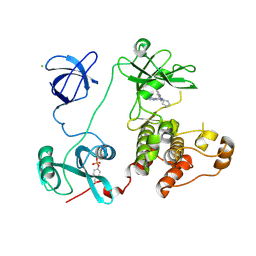 | | Crystal structure of HCK complexed with a pyrazolo-pyrimidine inhibitor 1-cyclopentyl-3-(1H-pyrrolo[2,3-b]pyridin-5-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine | | Descriptor: | 1-cyclopentyl-3-(1H-pyrrolo[2,3-b]pyridin-5-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Honda, K, Niwa, H, Toyama, M, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VX8
 
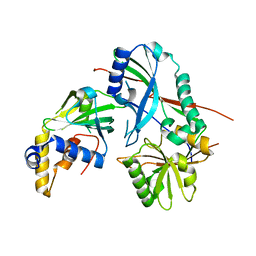 | |
3WPU
 
 | | Full-length beta-fructofuranosidase from Microbacterium saccharophilum K-1 | | Descriptor: | Beta-fructofuranosidase, GLYCEROL | | Authors: | Yokoi, G, Mori, M, Sato, S, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-01-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhancing thermostability and the structural characterization of Microbacterium saccharophilum K-1 beta-fructofuranosidase
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3WPV
 
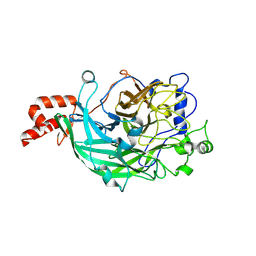 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase mutant T47S/F447V/F470Y/P500S | | Descriptor: | Beta-fructofuranosidase, GLYCEROL | | Authors: | Yokoi, G, Mori, M, Sato, S, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-01-17 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Enhancing thermostability and the structural characterization of Microbacterium saccharophilum K-1 beta-fructofuranosidase
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3WDP
 
 | | Structural analysis of a beta-glucosidase mutant derived from a hyperthermophilic tetrameric structure | | Descriptor: | Beta-glucosidase, GLYCEROL, PHOSPHATE ION | | Authors: | Nakabayashi, M, Kataoka, M, Ishikawa, K. | | Deposit date: | 2013-06-19 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of beta-glucosidase mutants derived from a hyperthermophilic tetrameric structure.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WPY
 
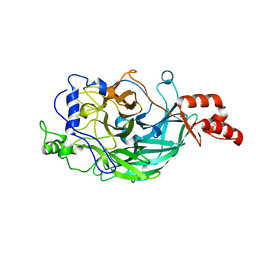 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase mutant T47S/S200T/F447V/P500S | | Descriptor: | Beta-fructofuranosidase | | Authors: | Yokoi, G, Mori, M, Sato, S, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-01-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enhancing thermostability and the structural characterization of Microbacterium saccharophilum K-1 beta-fructofuranosidase
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3WPZ
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase mutant T47S/S200T/F447P/F470Y/P500S | | Descriptor: | Beta-fructofuranosidase | | Authors: | Yokoi, G, Mori, M, Sato, S, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-01-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Enhancing thermostability and the structural characterization of Microbacterium saccharophilum K-1 beta-fructofuranosidase
Appl.Microbiol.Biotechnol., 98, 2014
|
|
2F7E
 
 | | PKA complexed with (S)-2-(1H-Indol-3-yl)-1-(5-isoquinolin-6-yl-pyridin-3-yloxymethyl-etylamine | | Descriptor: | (1S)-2-(1H-INDOL-3-YL)-1-{[(5-ISOQUINOLIN-6-YLPYRIDIN-3-YL)OXY]METHYL}ETHYLAMINE, PKI inhibitory peptide, cAMP-dependent protein kinase, ... | | Authors: | Stoll, V.S. | | Deposit date: | 2005-11-30 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structure-activity relationship of 3,4'-bispyridinylethylenes: discovery of a potent 3-isoquinolinylpyridine inhibitor of protein kinase B (PKB/Akt) for the treatment of cancer.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2GHG
 
 | | h-CHK1 complexed with A431994 | | Descriptor: | 5-{5-[(S)-2-AMINO-3-(1H-INDOL-3-YL)-PROPOXYL]-PYRIDIN-3-YL}-3-[1-(1H-PYRROL-2-YL)-METH-(Z)-YLIDENE]-1,3-DIHYDRO-INDOL-2-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2006-03-27 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery and SAR of oxindole-pyridine-based protein kinase B/Akt inhibitors for treating cancers.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5X9O
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1a | | Descriptor: | 1,2-ETHANEDIOL, 5-[(2-chloranyl-4-nitro-phenyl)amino]-1,3-dihydrobenzimidazol-2-one, B-cell lymphoma 6 protein, ... | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
2D8B
 
 | | Solution structure of the second tandem cofilin-domain of mouse twinfilin | | Descriptor: | Twinfilin-1 | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Sato, M, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-02 | | Release date: | 2006-06-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains
Protein Sci., 18, 2009
|
|
5YBX
 
 | | Crystal structure of the N-terminal domain of Bqt4 in S.pombe | | Descriptor: | Bouquet formation protein 4 | | Authors: | Hu, C, Chen, Y. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-19 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | The Inner Nuclear Membrane Protein Bqt4 in Fission Yeast Contains a DNA-Binding Domain Essential for Telomere Association with the Nuclear Envelope.
Structure, 27, 2019
|
|
5YC2
 
 | |
5YCA
 
 | | Crystal structure of inner membrane protein Bqt4 in complex with LEM2 | | Descriptor: | Lap-Emerin-Man domain protein 2, Ubiquitin-like protein SMT3,Bouquet formation protein 4 | | Authors: | Chen, Y, Hu, C. | | Deposit date: | 2017-09-07 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural insights into chromosome attachment to the nuclear envelope by an inner nuclear membrane protein Bqt4 in fission yeast.
Nucleic Acids Res., 47, 2019
|
|
5ZLH
 
 | | Crystal structure of Mn-ProtoporphyrinIX-reconstituted P450BM3 | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, MANGANESE PROTOPORPHYRIN IX | | Authors: | Omura, K, Aiba, Y, Onoda, H, Sugimoto, H, Shoji, O, Watanabe, Y. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Reconstitution of full-length P450BM3 with an artificial metal complex by utilising the transpeptidase Sortase A.
Chem. Commun. (Camb.), 54, 2018
|
|
