7XYD
 
 | | Crystal structure of TMPRSS2 in complex with Nafamostat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Liu, X, Duan, Y, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-06-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
7Y0F
 
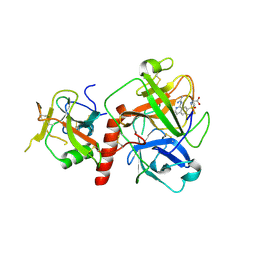 | | Crystal structure of TMPRSS2 in complex with UK-371804 | | Descriptor: | 2-[(1-carbamimidamido-4-chloranyl-isoquinolin-7-yl)sulfonylamino]-2-methyl-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, H, Duan, Y, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-06-04 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
7Y0E
 
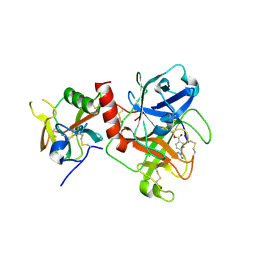 | | Crystal structure of TMPRSS2 in complex with Camostat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Duan, Y, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-06-04 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
6WMW
 
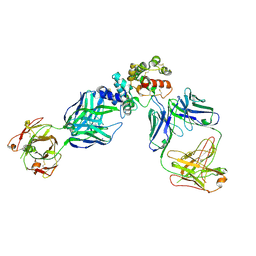 | | GFRAL receptor bound with two antibody Fabs (3P10, 25M22) | | Descriptor: | FAB25M22 heavy chain fragment, FAB25M22 light chain, FAB3P10 heavy chain fragment, ... | | Authors: | White, A, Lakshminarasimhan, D, Olland, A, Suto, R.K. | | Deposit date: | 2020-04-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Antibody-mediated inhibition of GDF15-GFRAL activity reverses cancer cachexia in mice.
Nat Med, 26, 2020
|
|
7T9U
 
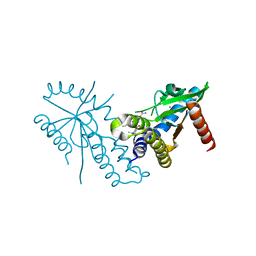 | | Crystal structure of hSTING with an agonist (SHR169224) | | Descriptor: | (3S,4S)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4-(prop-2-en-1-yl)-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
7T9V
 
 | | Crystal structure of hSTING with the agonist, SHR171032 | | Descriptor: | (3S,4S)-4-(3-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methoxy-1H-benzimidazol-1-yl}propyl)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
6KG6
 
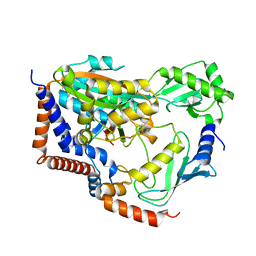 | | Crystal structure of MavC/UBE2N-Ub complex | | Descriptor: | MavC, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 N | | Authors: | Wang, Y, Huang, Y, Chang, M, Feng, Y. | | Deposit date: | 2019-07-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC.
Nat Commun, 11, 2020
|
|
7YV8
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike glycoprotein, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7YVU
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with mouse ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-08-19 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
6KYF
 
 | | Crystal structure of an anti-CRISPR protein | | Descriptor: | AcrF11, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Niu, Y, Wang, H, Zhang, Y, Feng, Y. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | A Type I-F Anti-CRISPR Protein Inhibits the CRISPR-Cas Surveillance Complex by ADP-Ribosylation.
Mol.Cell, 80, 2020
|
|
7V1N
 
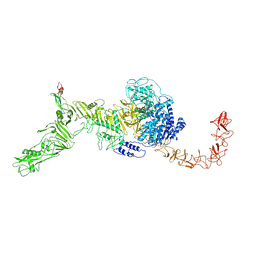 | | Structure of the Clade 2 C. difficile TcdB in complex with its receptor TFPI | | Descriptor: | Isoform Beta of Tissue factor pathway inhibitor, Toxin B | | Authors: | Luo, J, Yang, Q, Zhang, X, Zhang, Y, Wan, L, Li, Y, Tao, L. | | Deposit date: | 2021-08-05 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | TFPI is a colonic crypt receptor for TcdB from hypervirulent clade 2 C. difficile.
Cell, 185, 2022
|
|
5KZF
 
 | |
4ZS6
 
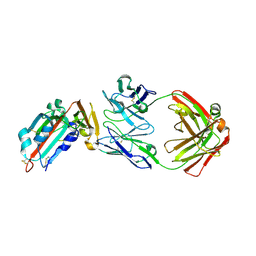 | | Receptor binding domain and Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein, fab Heavy Chain, ... | | Authors: | Yu, X, Wang, X. | | Deposit date: | 2015-05-13 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.166 Å) | | Cite: | Structural basis for the neutralization of MERS-CoV by a human monoclonal antibody MERS-27
Sci Rep, 5, 2015
|
|
7CKI
 
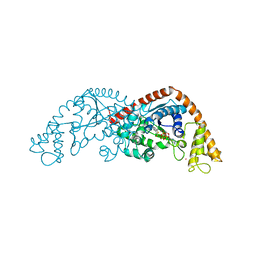 | | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with chuangxinmycin and ATP | | Descriptor: | (5~{S},6~{R})-5-methyl-7-thia-2-azatricyclo[6.3.1.0^{4,12}]dodeca-1(12),3,8,10-tetraene-6-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lyu, G, Fan, S, Jin, Y, Liu, J, Zou, S, Wu, G, Yang, Z. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with chuangxinmycin and ATP
To Be Published
|
|
4WAU
 
 | | Crystal structure of CENP-M solved by native-SAD phasing | | Descriptor: | Centromere protein M | | Authors: | Weinert, T, Basilico, F, Cecatiello, V, Pasqualato, S, Wang, M. | | Deposit date: | 2014-09-01 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
5KWA
 
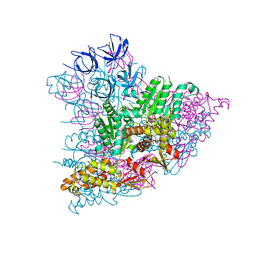 | |
7BZ5
 
 | | Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of B38, Light chain of B38, ... | | Authors: | Wu, Y, Qi, J, Gao, F. | | Deposit date: | 2020-04-26 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2.
Science, 368, 2020
|
|
3AFK
 
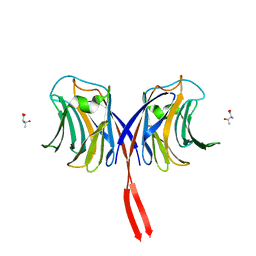 | |
5F6L
 
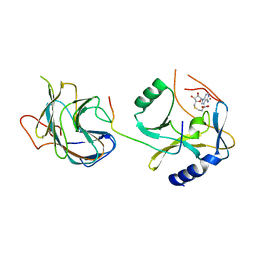 | | The crystal structure of MLL1 (N3861I/Q3867L) in complex with RbBP5 and Ash2L | | Descriptor: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-06 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
5F5E
 
 | |
5F6K
 
 | | Crystal structure of the MLL3-Ash2L-RbBP5 complex | | Descriptor: | Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-06 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
8GRY
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with rat ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-09-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
5F59
 
 | | The crystal structure of MLL3 SET domain | | Descriptor: | Histone-lysine N-methyltransferase 2C, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-04 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
6CWA
 
 | |
6KXW
 
 | | Crystal structure of human aquaporin AQP7 in bound to glycerol | | Descriptor: | Aquaporin-7, GLYCEROL | | Authors: | Zhang, L, Yao, D, Zhou, F, Zhang, Q, Zhou, L, Cao, Y. | | Deposit date: | 2019-09-13 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The structural basis for glycerol permeation by human AQP7
Sci Bull (Beijing), 66, 2020
|
|
