2IPA
 
 | | solution structure of Trx-ArsC complex | | Descriptor: | Protein arsC, Thioredoxin | | Authors: | Jin, C, Hu, Y, Li, Y, Zhang, X. | | Deposit date: | 2006-10-12 | | Release date: | 2007-02-13 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Conformational fluctuations coupled to the thiol-disulfide transfer between thioredoxin and arsenate reductase in Bacillus subtilis.
J.Biol.Chem., 282, 2007
|
|
6D2K
 
 | | Crystal structure of the FERM domain of mouse FARP2 | | Descriptor: | FERM, ARHGEF and pleckstrin domain-containing protein 2 | | Authors: | He, X, Zhang, X. | | Deposit date: | 2018-04-13 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analyses of FERM domain-mediated membrane localization of FARP1.
Sci Rep, 8, 2018
|
|
6D21
 
 | |
6NT9
 
 | | Cryo-EM structure of the complex between human TBK1 and chicken STING | | Descriptor: | Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of STING binding with and phosphorylation by TBK1.
Nature, 567, 2019
|
|
4M8M
 
 | |
4MGG
 
 | | Crystal structure of an enolase (mandelate racemase subgroup) from labrenzia aggregata iam 12614 (target nysgrc-012903) with bound mg, space group p212121 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Muconate lactonizing enzyme, ... | | Authors: | Vetting, M.W, Zhang, X, Wasserman, S.R, Morisco, L.L, Sojitra, S, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup) from labrenzia aggregata iam 12614 (target nysgrc-012903) with bound mg, space group p212121
To be Published
|
|
8Y89
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 2TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Wang, H.F, Zhang, X, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8A
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 3TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8E
 
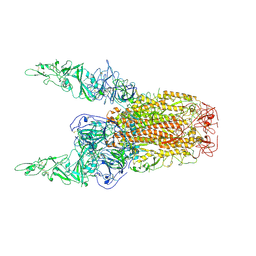 | | Structure of HCoV-HKU1C spike in the inactive-2up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y88
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-2up conformation with 2TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y7X
 
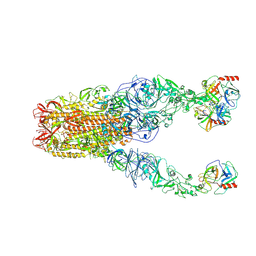 | | Structure of HCoV-HKU1A spike in the functionally anchored-3up conformation with 3TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8D
 
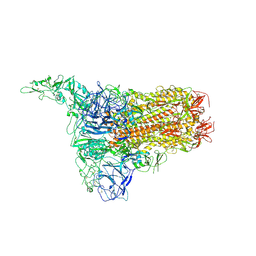 | | Structure of HCoV-HKU1C spike in the inactive-1up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y7Y
 
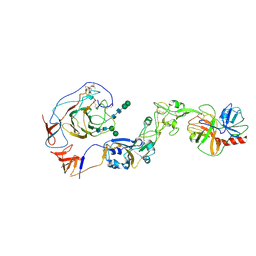 | | Local structure of HCoV-HKU1A spike in complex with TMPRSS2 and glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, H.F, Zhang, X, Lu, Y, Liu, X, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8F
 
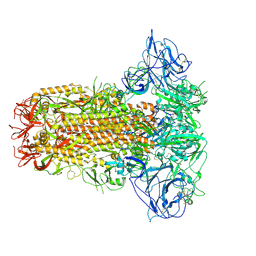 | | Structure of HCoV-HKU1C spike in the glycan-activated-closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8I
 
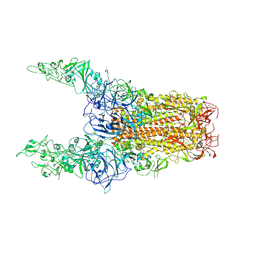 | | Structure of HCoV-HKU1C spike in the glycan-activated-3up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8J
 
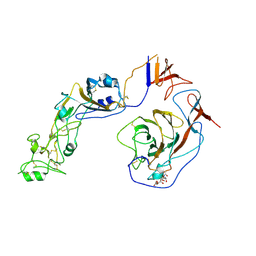 | | Local structure of HCoV-HKU1C spike in complex with glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8C
 
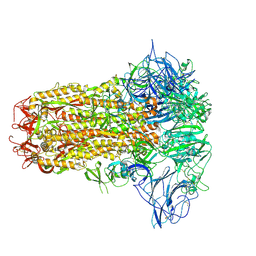 | | Structure of HCoV-HKU1C spike in the inactive-closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y87
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-1up conformation with 1TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8G
 
 | | Structure of HCoV-HKU1C spike in the glycan-activated-1up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8B
 
 | | Local structure of HCoV-HKU1C spike in complex with TMPRSS2 and glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, H.F, Zhang, X, Lu, Y.C, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8H
 
 | | Structure of HCoV-HKU1C spike in the glycan-activated-2up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
6S8H
 
 | | Cryo-EM structure of LptB2FG in complex with LPS | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
6S8N
 
 | | Cryo-EM structure of LptB2FGC in complex with lipopolysaccharide | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, Inner membrane protein yjgQ, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
6S8G
 
 | | Cryo-EM structure of LptB2FGC in complex with AMP-PNP | | Descriptor: | Inner membrane protein yjgQ, LPS export ABC transporter permease LptF, Lipopolysaccharide ABC transporter, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
6NT5
 
 | | Cryo-EM structure of full-length human STING in the apo state | | Descriptor: | Stimulator of interferon protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
