8AQ2
 
 | | In meso structure of the membrane integral lipoprotein N-acyltransferase Lnt from P. aeruginosa covalently linked with TITC | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8AQ3
 
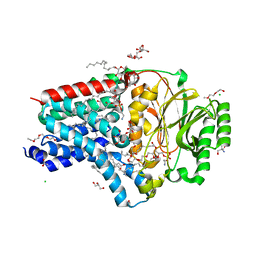 | | In surfo structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli in complex with PE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8AQ4
 
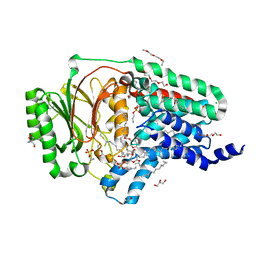 | | In surfo structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli in complex with TITC and lyso-PE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
5FKJ
 
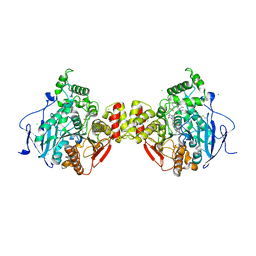 | | Crystal structure of mouse acetylcholinesterase in complex with C-547, an alkyl ammonium derivative of 6-methyl uracil | | Descriptor: | 1,3-BIS[5(DIETHYL-O-NITROBENZYLAMMONIUM)PENTYL]-6-METHYLURACIL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Nachon, F, Villard-Wandhammer, M, Petrov, K, Masson, P. | | Deposit date: | 2015-10-16 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.133 Å) | | Cite: | Slow-Binding Inhibition of Acetylcholinesterase by a 6-Methyluracil Alkyl-Ammonium Derivative: Mechanism and Advantages for Myasthenia Gravis Treatment.
Biochem.J., 473, 2016
|
|
3QZQ
 
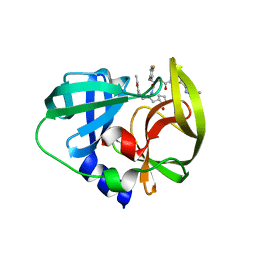 | | Human enterovirus 71 3C protease mutant E71D in complex with rupintrivir | | Descriptor: | 3C protein, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Wang, J, Fan, T, Yao, X, Wu, Z, Guo, L, Lei, X, Wang, J, Wang, M, Jin, Q, Cui, S. | | Deposit date: | 2011-03-07 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7001 Å) | | Cite: | Crystal Structures of Enterovirus 71 3C Protease Complexed with Rupintrivir Reveal the Roles of Catalytically Important Residues.
J.Virol., 85, 2011
|
|
7NKW
 
 | | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Engilberge, S, Huang, C.-Y, Leonarski, F, Wojdyla, J.A, Marsh, M, Olieric, V, Wang, M. | | Deposit date: | 2021-02-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector
To Be Published
|
|
3R0F
 
 | | Human enterovirus 71 3C protease mutant H133G in complex with rupintrivir | | Descriptor: | 1,2-ETHANEDIOL, 3C protein, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Wang, J, Fan, T, Yao, X, Wu, Z, Guo, L, Lei, X, Wang, J, Wang, M, Jin, Q, Cui, S. | | Deposit date: | 2011-03-08 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3083 Å) | | Cite: | Crystal Structures of Enterovirus 71 3C Protease Complexed with Rupintrivir Reveal the Roles of Catalytically Important Residues.
J.Virol., 85, 2011
|
|
4K48
 
 | | Structure of the Streptococcus pneumoniae leucyl-tRNA synthetase editing domain | | Descriptor: | Leucine--tRNA ligase | | Authors: | Hu, Q.H, Liu, R.J, Fang, Z.P, Zhang, J, Ding, Y.Y, Tan, M, Wang, M, Pan, W, Zhou, H.C, Wang, E.D. | | Deposit date: | 2013-04-12 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery of a potent benzoxaborole-based anti-pneumococcal agent targeting leucyl-tRNA synthetase
Sci Rep, 3, 2013
|
|
4AYL
 
 | | Molecular structure of a metal-independent bacterial glycosyltransferase that catalyzes the synthesis of histo-blood group A antigen | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BOGT-METAL-INDEPENDENT GLYCOSYLTRANSFERASE, CALCIUM ION, ... | | Authors: | Thiyagarajan, N, Pham, T.T.K, Stinsonb, B, Sundriyala, A, Tumbale, P, Lizotte-Waniewskib, M, Brewb, K, Acharya, K.R. | | Deposit date: | 2012-06-21 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Structure of a Metal-Independent Bacterial Glycosyltransferase that Catalyzes the Synthesis of Histo-Blood Group a Antigen
Sci.Rep., 2, 2012
|
|
4AYJ
 
 | | Molecular structure of a metal-independent bacterial glycosyltransferase that catalyzes the synthesis of histo-blood group A antigen | | Descriptor: | BOGT - METAL-INDEPENDENT GLYCOSYLTRANSFERASE, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Thiyagarajan, N, Pham, T.T.K, Stinson, B, Sundriyal, A, Tumbale, P, Lizotte-Waniewskib, M, Brewb, K, Acharya, K.R. | | Deposit date: | 2012-06-21 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a Metal-Independent Bacterial Glycosyltransferase that Catalyzes the Synthesis of Histo-Blood Group a Antigen
Sci.Rep., 2, 2012
|
|
3QZR
 
 | | Human enterovirus 71 3C protease mutant E71A in complex with rupintrivir | | Descriptor: | 1,2-ETHANEDIOL, 3C protein, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Wang, J, Fan, T, Yao, X, Wu, Z, Guo, L, Lei, X, Wang, J, Wang, M, Jin, Q, Cui, S. | | Deposit date: | 2011-03-07 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.039 Å) | | Cite: | Crystal Structures of Enterovirus 71 3C Protease Complexed with Rupintrivir Reveal the Roles of Catalytically Important Residues.
J.Virol., 85, 2011
|
|
4KUD
 
 | | Crystal structure of N-terminal acetylated Sir3 BAH domain D205N mutant in complex with yeast nucleosome core particle | | Descriptor: | Histone H2A.2, Histone H2B.1, Histone H3, ... | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KUI
 
 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain | | Descriptor: | ACETIC ACID, ISOPROPYL ALCOHOL, Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4K47
 
 | | Structure of the Streptococcus pneumoniae leucyl-tRNA synthetase editing domain bound to a benzoxaborole-AMP adduct | | Descriptor: | Leucine--tRNA ligase, SULFATE ION, [(1R,5R,6R,8S)-6-(6-aminopurin-9-yl)-3'-[(R)-oxidanyl(phenyl)methyl]spiro[2,4,7-trioxa-3-boranuidabicyclo[3.3.0]octane-3,9'-8-oxa-9-boranuidabicyclo[4.3.0]nona-1,3,5-triene]-8-yl]methyl dihydrogen phosphate | | Authors: | Hu, Q.H, Liu, R.J, Fang, Z.P, Zhang, J, Ding, Y.Y, Tan, M, Wang, M, Pan, W, Zhou, H.C, Wang, E.D. | | Deposit date: | 2013-04-12 | | Release date: | 2013-09-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery of a potent benzoxaborole-based anti-pneumococcal agent targeting leucyl-tRNA synthetase
Sci Rep, 3, 2013
|
|
1Y0H
 
 | | Structure of Rv0793 from Mycobacterium tuberculosis | | Descriptor: | ACETATE ION, hypothetical protein Rv0793 | | Authors: | Lemieux, M.J, Ference, C, Cherney, M.M, Wang, M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-11-15 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Rv0793, a hypothetical monooxygenase from M. tuberculosis
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
4KUL
 
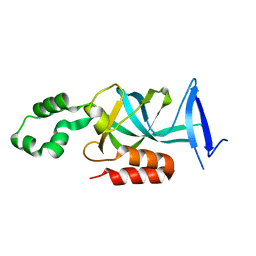 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain V83P mutant | | Descriptor: | Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3ZJ7
 
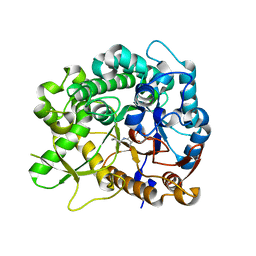 | | Crystal structure of strictosidine glucosidase in complex with inhibitor-1 | | Descriptor: | (1R,2S,3S,4R,5R)-4-(cyclohexylamino)-5-(hydroxymethyl)cyclopentane-1,2,3-triol, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
6OM3
 
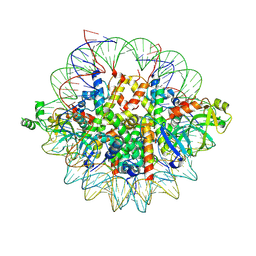 | | Crystal structure of the Orc1 BAH domain in complex with a nucleosome core particle | | Descriptor: | DNA (146-MER), DNA (147-MER), Histone H2A, ... | | Authors: | De Ioannes, P.E, Wang, M, Armache, K.-J. | | Deposit date: | 2019-04-18 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and function of the Orc1 BAH-nucleosome complex.
Nat Commun, 10, 2019
|
|
2PQE
 
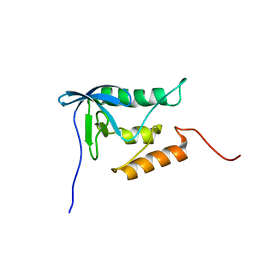 | | Solution structure of proline-free mutant of staphylococcal nuclease | | Descriptor: | Thermonuclease | | Authors: | Shan, L, Tong, Y, Xie, T, Wang, M, Wang, J. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Restricted backbone conformational and motional flexibilities of loops containing peptidyl-proline bonds dominate the enzyme activity of staphylococcal nuclease.
Biochemistry, 46, 2007
|
|
3OSY
 
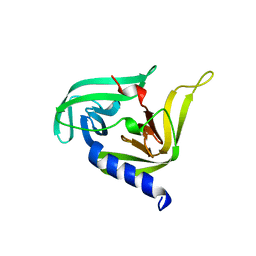 | |
3ZJ8
 
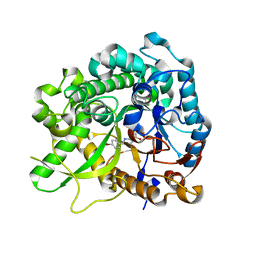 | | Crystal structure of strictosidine glucosidase in complex with inhibitor-2 | | Descriptor: | (1R,2S,3S,4R,5R)-4-[(4-bromophenyl)methylamino]-5-(hydroxymethyl)cyclopentane-1,2,3-triol, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
5DWK
 
 | | Diacylglycerol Kinase solved by multi crystal multi orientation native SAD | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, ACETATE ION, ... | | Authors: | Weinert, T, Olieric, V, Finke, A.D, Li, D, Caffrey, M, Wang, M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Data-collection strategy for challenging native SAD phasing.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4A3Y
 
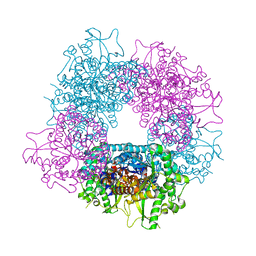 | | Crystal structure of Raucaffricine glucosidase from ajmaline biosynthesis pathway | | Descriptor: | GLYCEROL, RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, SULFATE ION | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Barleben, L, Rajendran, C, Lin, H, Stoeckigt, J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity.
Acs Chem.Biol., 7, 2012
|
|
6JVX
 
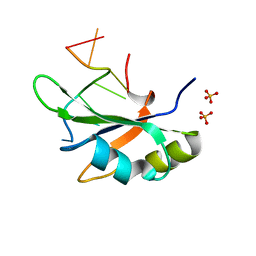 | | Crystal structure of RBM38 in complex with RNA | | Descriptor: | RNA (5'-R(*UP*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*G)-3'), RNA-binding protein 38, SULFATE ION | | Authors: | Qian, K, Li, M, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2019-04-17 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis for mRNA recognition by human RBM38.
Biochem.J., 477, 2020
|
|
4PGO
 
 | | Crystal structure of hypothetical protein PF0907 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | CHLORIDE ION, Uncharacterized protein | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-05-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
