7RE2
 
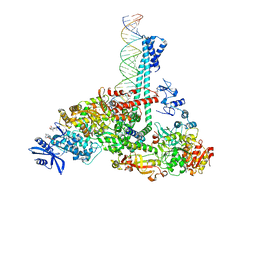 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(1)-RTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RDY
 
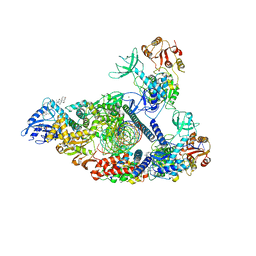 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - engaged class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE0
 
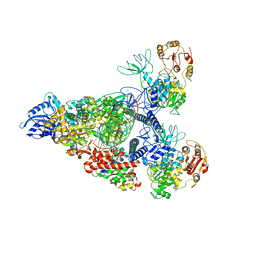 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - swiveled class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7A5P
 
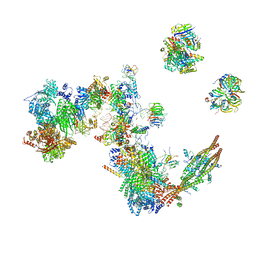 | | Human C Complex Spliceosome - Medium-resolution PERIPHERY | | Descriptor: | Cell division cycle 5-like protein, Crooked neck-like protein 1, Eukaryotic initiation factor 4A-III, ... | | Authors: | Bertram, K, Kastner, B. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural Insights into the Roles of Metazoan-Specific Splicing Factors in the Human Step 1 Spliceosome.
Mol.Cell, 80, 2020
|
|
6Y53
 
 | | human 17S U2 snRNP low resolution part | | Descriptor: | HIV Tat-specific factor 1, Probable ATP-dependent RNA helicase DDX46, Small nuclear ribonucleoprotein E, ... | | Authors: | Zhang, Z, Will, C.L, Bertram, K, Luehrmann, R, Stark, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Molecular architecture of the human 17S U2 snRNP.
Nature, 583, 2020
|
|
7RDX
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - open class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6ZYM
 
 | | Human C Complex Spliceosome - High-resolution CORE | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Corepressor interacting with RBPJ 1, ... | | Authors: | Bertram, K, Kastner, B. | | Deposit date: | 2020-08-02 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insights into the Roles of Metazoan-Specific Splicing Factors in the Human Step 1 Spliceosome.
Mol.Cell, 80, 2020
|
|
5UAE
 
 | | Crystal structure of the coiled-coil domain from Listeria Innocua Phage Integrase (Trigonal Form) | | Descriptor: | CITRATE ANION, Putative integrase | | Authors: | Gupta, K, Sharp, R, Yuan, J.B, Van Duyne, G.D. | | Deposit date: | 2016-12-19 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Coiled-coil interactions mediate serine integrase directionality.
Nucleic Acids Res., 45, 2017
|
|
5UDO
 
 | |
8ELG
 
 | |
8ELH
 
 | |
5U96
 
 | |
7AYY
 
 | | Structure of the human 8-oxoguanine DNA Glycosylase hOGG1 in complex with activator TH10785 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Davies, J.R, Stenmark, P. | | Deposit date: | 2020-11-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-molecule activation of OGG1 increases oxidative DNA damage repair by gaining a new function.
Science, 376, 2022
|
|
7AYZ
 
 | |
7AZ0
 
 | |
7TUX
 
 | | Crystal Structure of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase in complex with [(3S)-4-Hydroxy-3-[({2-amino-4-hydroxy-5H-pyrrolo[3,2-d]pyrimidin-7-yl}methyl)amino]butyl]phosphonic acid | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Hypoxanthine-guanine-xanthine phosphoribosyltransferase, ... | | Authors: | Harijan, R.K, Minnow, Y.V.T, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2022-02-03 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inhibition and Mechanism of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase.
Acs Chem.Biol., 17, 2022
|
|
3DXC
 
 | |
3DXD
 
 | |
7WVL
 
 | |
3DXE
 
 | |
3D8F
 
 | | Crystal structure of the human Fe65-PTB1 domain with bound phosphate (trigonal crystal form) | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1, PHOSPHATE ION | | Authors: | Radzimanowski, J, Ravaud, S, Sinning, I, Wild, K. | | Deposit date: | 2008-05-23 | | Release date: | 2008-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the human Fe65-PTB1 domain.
J.Biol.Chem., 283, 2008
|
|
3D8D
 
 | | Crystal structure of the human Fe65-PTB1 domain | | Descriptor: | 1,2-ETHANEDIOL, Amyloid beta A4 precursor protein-binding family B member 1, MERCURY (II) ION | | Authors: | Radzimanowski, J, Ravaud, S, Sinning, I, Wild, K. | | Deposit date: | 2008-05-23 | | Release date: | 2008-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human Fe65-PTB1 domain.
J.Biol.Chem., 283, 2008
|
|
1CXR
 
 | |
7KMG
 
 | | LY-CoV555 neutralizing antibody against SARS-CoV-2 | | Descriptor: | GLYCEROL, LY-CoV555 Fab heavy chain, LY-CoV555 Fab light chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
7KMH
 
 | | LY-CoV488 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LY-CoV488 Fab heavy chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Boyles, J.S, Dickinson, C.D, Coleman, K.A. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
