3ZXH
 
 | |
7LJE
 
 | | Discovery of Spirohydantoins as Selective, Orally Bioavailable Inhibitors of p300/CBP Histone Acetyltransferases | | Descriptor: | 2-[4-[(3'R,4S)-3'-fluoro-1-[2-[(4-fluorophenyl)methyl-[(1S)-2,2,2-trifluoro-1-methyl-ethyl]amino]-2-oxo-ethyl]-2,5-dioxo-spiro[imidazolidine-4,1'-indane]-5'-yl]pyrazol-1-yl]-N-methyl-acetamide, Histone acetyltransferase p300 | | Authors: | Jakob, C.G. | | Deposit date: | 2021-01-29 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.607 Å) | | Cite: | Discovery of spirohydantoins as selective, orally bioavailable inhibitors of p300/CBP histone acetyltransferases.
Bioorg.Med.Chem.Lett., 39, 2021
|
|
9GSS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH S-HEXYL GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE P1-1, S-HEXYLGLUTATHIONE, ... | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
8GSS
 
 | | Human glutathione S-transferase P1-1, complex with glutathione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1, ... | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
3ELM
 
 | | Crystal Structure of MMP-13 Complexed with Inhibitor 24f | | Descriptor: | (2R)-({[5-(4-ethoxyphenyl)thiophen-2-yl]sulfonyl}amino){1-[(1-methylethoxy)carbonyl]piperidin-4-yl}ethanoic acid, CALCIUM ION, Collagenase 3, ... | | Authors: | Kulathila, R, Monovich, L, Koehn, J. | | Deposit date: | 2008-09-22 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of potent, selective, and orally active carboxylic acid based inhibitors of matrix metalloproteinase-13
J.Med.Chem., 52, 2009
|
|
4ZVV
 
 | | Lactate dehydrogenase A in complex with a trisubstituted piperidine-2,4-dione inhibitor GNE-140 | | Descriptor: | (2~{R})-5-(2-chlorophenyl)sulfanyl-2-(4-morpholin-4-ylphenyl)-4-oxidanyl-2-thiophen-3-yl-1,3-dihydropyridin-6-one, L-lactate dehydrogenase A chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Li, Y, Chen, Z, Eigenbrot, C. | | Deposit date: | 2015-05-18 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Metabolic plasticity underpins innate and acquired resistance to LDHA inhibition.
Nat.Chem.Biol., 12, 2016
|
|
3PUP
 
 | | Structure of Glycogen Synthase Kinase 3 beta (GSK3B) in complex with a ruthenium octasporine ligand (OS1) | | Descriptor: | Glycogen synthase kinase-3 beta, Ruthenium octasporine | | Authors: | Filippakopoulos, P, Kraling, K, Essen, L.O, Meggers, E, Knapp, S. | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structurally sophisticated octahedral metal complexes as highly selective protein kinase inhibitors.
J.Am.Chem.Soc., 133, 2011
|
|
6R99
 
 | |
1BD8
 
 | | STRUCTURE OF CDK INHIBITOR P19INK4D | | Descriptor: | P19INK4D CDK4/6 INHIBITOR | | Authors: | Baumgartner, R, Fernandez-Catalan, C, Winoto, A, Huber, R, Engh, R, Holak, T.A. | | Deposit date: | 1998-05-12 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human cyclin-dependent kinase inhibitor p19INK4d: comparison to known ankyrin-repeat-containing structures and implications for the dysfunction of tumor suppressor p16INK4a.
Structure, 6, 1998
|
|
1BIP
 
 | |
8G5C
 
 | |
8GHN
 
 | |
8GHL
 
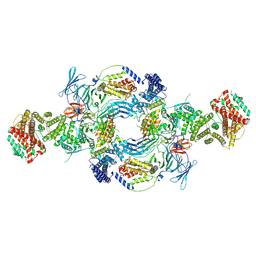 | | the Hir complex core | | Descriptor: | Histone transcription regulator 3, Protein HIR1, Protein HIR2 | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-10 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 2024
|
|
8GHA
 
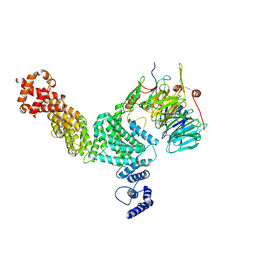 | | Hir3 Arm/Tail, Hir2 WD40, C-terminal Hpc2 | | Descriptor: | Histone promoter control protein 2, Histone transcription regulator 3, Protein HIR2 | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 2024
|
|
8GHM
 
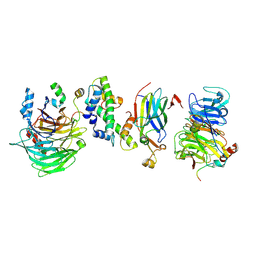 | | Hir1 WD40 domains and Asf1/H3/H4 | | Descriptor: | Histone H3, Histone H4, Histone chaperone, ... | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 2024
|
|
4KVO
 
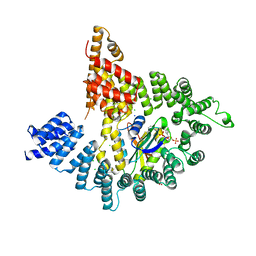 | |
4KVM
 
 | |
4KVX
 
 | | Crystal structure of Naa10 (Ard1) bound to AcCoA | | Descriptor: | ACETYL COENZYME *A, N-terminal acetyltransferase A complex catalytic subunit ard1 | | Authors: | Liszczak, G.P, Marmorstein, R.Q. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for N-terminal acetylation by the heterodimeric NatA complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
5CMC
 
 | |
5CMB
 
 | |
8V0F
 
 | |
4PGT
 
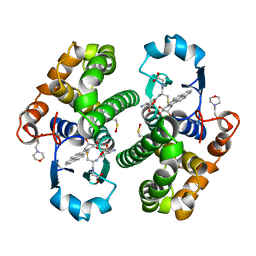 | | CRYSTAL STRUCTURE OF HGSTP1-1[V104] COMPLEXED WITH THE GSH CONJUGATE OF (+)-ANTI-BPDE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-4-[1-(CARBOXYMETHYL-CARBAMOYL)-2-(9-HYDROXY-7,8-DIOXO-7,8,9,10-TETRAHYDRO-BENZO[DEF]CHRYSEN-10-YLSULFANYL)-ETHYLCARBAMOYL]-BUTYRIC ACID, PROTEIN (GLUTATHIONE S-TRANSFERASE), ... | | Authors: | Ji, X, Blaszczyk, J. | | Deposit date: | 1999-03-22 | | Release date: | 1999-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of residue 104 and water molecules in the xenobiotic substrate-binding site in human glutathione S-transferase P1-1.
Biochemistry, 38, 1999
|
|
5V5X
 
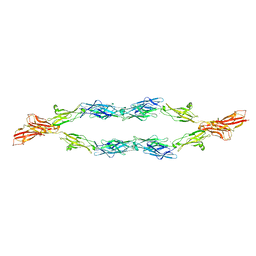 | | Protocadherin gammaB7 EC3-6 cis-dimer structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2017-03-15 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Protocadherin cis-dimer architecture and recognition unit diversity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2GSR
 
 | |
5YJE
 
 | | Crystal structure of HIRA(644-1017) | | Descriptor: | Protein HIRA, SULFATE ION | | Authors: | Sato, Y, Senda, M, Senda, T. | | Deposit date: | 2017-10-10 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional activity of the H3.3 histone chaperone complex HIRA requires trimerization of the HIRA subunit
Nat Commun, 9, 2018
|
|
