3OE9
 
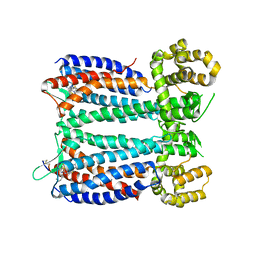 | | Crystal structure of the chemokine CXCR4 receptor in complex with a small molecule antagonist IT1t in P1 spacegroup | | Descriptor: | (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, Lysozyme Chimera | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3ODU
 
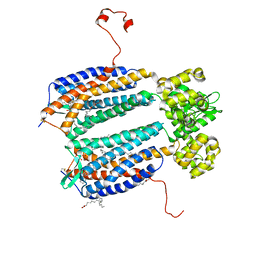 | | The 2.5 A structure of the CXCR4 chemokine receptor in complex with small molecule antagonist IT1t | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3OE6
 
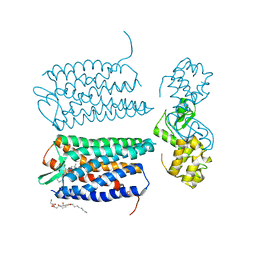 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in I222 spacegroup | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3OE8
 
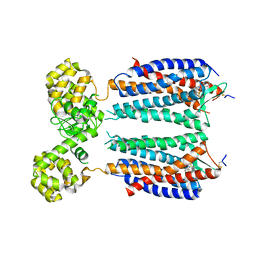 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in P1 spacegroup | | Descriptor: | (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, Lysozyme Chimera | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3RZE
 
 | | Structure of the human histamine H1 receptor in complex with doxepin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (3E)-3-(dibenzo[b,e]oxepin-11(6H)-ylidene)-N,N-dimethylpropan-1-amine, (3Z)-3-(dibenzo[b,e]oxepin-11(6H)-ylidene)-N,N-dimethylpropan-1-amine, ... | | Authors: | Shimamura, T, Han, G.W, Shiroishi, M, Weyand, S, Tsujimoto, H, Winter, G, Katritch, V, Abagyan, R, Cherezov, V, Liu, W, Kobayashi, T, Stevens, R, Iwata, S, GPCR Network (GPCR) | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the human histamine H1 receptor complex with doxepin.
Nature, 475, 2011
|
|
2H1Y
 
 | |
2GLM
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) from Helicobacter pylori complexed with Compound 2 | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 2-CHLORO-5-(5-{(E)-[(2Z)-3-(2-METHOXYETHYL)-4-OXO-2-(PHENYLIMINO)-1,3-THIAZOLIDIN-5-YLIDENE]METHYL}-2-FURYL)BENZOIC ACID, BENZAMIDINE, ... | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
8TES
 
 | | Human cytomegalovirus portal vertex, virion configuration 2 (VC2) | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large structural phosphoprotein, ... | | Authors: | Jih, J, Liu, Y.T, Liu, W, Zhou, H. | | Deposit date: | 2023-07-06 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Sci Adv, 10, 2024
|
|
8TEW
 
 | | Human cytomegalovirus penton vertex, CVSC-bound configuration | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large structural phosphoprotein, ... | | Authors: | Jih, J, Liu, Y.T, Liu, W, Zhou, H. | | Deposit date: | 2023-07-07 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Sci Adv, 10, 2024
|
|
8TET
 
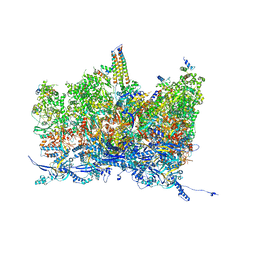 | | Human cytomegalovirus portal vertex, non-infectious enveloped particle (NIEP) configuration 1 (NC1) | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large structural phosphoprotein, ... | | Authors: | Jih, J, Liu, Y.T, Liu, W, Zhou, H. | | Deposit date: | 2023-07-07 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Sci Adv, 10, 2024
|
|
8TEP
 
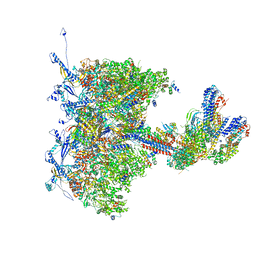 | | Human cytomegalovirus portal vertex, virion configuration 1 (VC1) | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Inner tegument protein, ... | | Authors: | Jih, J, Liu, Y.T, Liu, W, Zhou, H. | | Deposit date: | 2023-07-06 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Sci Adv, 10, 2024
|
|
8TEU
 
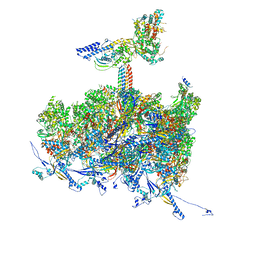 | | Human cytomegalovirus portal vertex, non-infectious enveloped particle (NIEP) configuration 2 - inverted (NC2-inv) | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large structural phosphoprotein, ... | | Authors: | Jih, J, Liu, Y.T, Liu, W, Zhou, H. | | Deposit date: | 2023-07-07 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Sci Adv, 10, 2024
|
|
2GLV
 
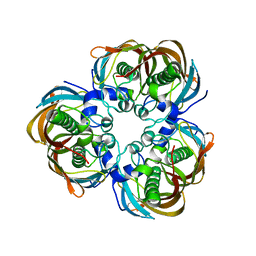 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) mutant(Y100A) from Helicobacter pylori | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, CHLORIDE ION | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
2GLL
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) from Helicobacter pylori | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, CHLORIDE ION | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
2GLP
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) from Helicobacter pylori complexed with compound 1 | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
5Y1Z
 
 | | Crystal structure of ZMYND8 PHD-BROMO-PWWP tandem in complex with Drebrin ADF-H domain | | Descriptor: | Drebrin, GLYCEROL, Protein kinase C-binding protein 1, ... | | Authors: | Yao, N, Li, J, Liu, H, Wan, J, Liu, W, Zhang, M. | | Deposit date: | 2017-07-22 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.676 Å) | | Cite: | The Structure of the ZMYND8/Drebrin Complex Suggests a Cytoplasmic Sequestering Mechanism of ZMYND8 by Drebrin
Structure, 25, 2017
|
|
5ZZ9
 
 | | Crystal structure of Homer2 EVH1/Drebrin PPXXF complex | | Descriptor: | Homer protein homolog 2, Peptide from Drebrin | | Authors: | Li, Z, Liu, H, Li, J, Liu, W, Zhang, M. | | Deposit date: | 2018-05-31 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Homer Tetramer Promotes Actin Bundling Activity of Drebrin.
Structure, 27, 2019
|
|
5YPI
 
 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EI1 complex | | Descriptor: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPK
 
 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EI2 complex | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-1, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPL
 
 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EP complex | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-1, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPN
 
 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem representing an EI2 complex | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Metallo-beta-lactamase NDM-1, SULFATE ION, ... | | Authors: | Feng, H, Liu, W, Wang, D. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPM
 
 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem representing an EI1 complex | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, Metallo-beta-lactamase NDM-1, SULFATE ION, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5HDN
 
 | |
5HDG
 
 | |
5HDK
 
 | |
