4R9C
 
 | |
4R9A
 
 | |
4R9B
 
 | |
4RL7
 
 | |
6G1I
 
 | | GH124 cellulase from Ruminiclostridium thermocellum in complex with Mn and fructosylated cellopentaose | | Descriptor: | Glycosyl Hydrolase, MALONIC ACID, MANGANESE (II) ION, ... | | Authors: | Urresti, S, Davies, G.J, Walton, P.H. | | Deposit date: | 2018-03-21 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structural studies of the unusual metal-ion site of the GH124 endoglucanase from Ruminiclostridium thermocellum.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6TN1
 
 | | Unliganded Crystal Structure of Recombinant GBA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FORMIC ACID, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2019-12-05 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | A baculoviral system for the production of human beta-glucocerebrosidase enables atomic resolution analysis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6G1G
 
 | |
6TJQ
 
 | | Crystal Structure of Recombinant GBA in Complex with 2-Deoxy-2-fluoro-beta-D-glucopyranoside | | Descriptor: | (2~{R},3~{S},4~{S},5~{S})-5-fluoranyl-2-(hydroxymethyl)oxane-3,4-diol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2019-11-26 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A baculoviral system for the production of human beta-glucocerebrosidase enables atomic resolution analysis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6TJK
 
 | | Crystal Structure of Recombinant GBA in Complex with Bis-Tris Propane. | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2019-11-26 | | Release date: | 2020-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A baculoviral system for the production of human beta-glucocerebrosidase enables atomic resolution analysis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6TJJ
 
 | | Structure of Cerezyme at pH 4.6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2019-11-26 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A baculoviral system for the production of human beta-glucocerebrosidase enables atomic resolution analysis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8ILQ
 
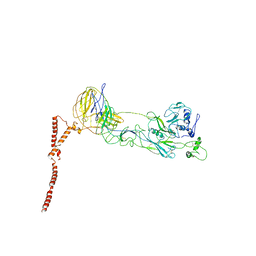 | | Structure of SFTSV Gn-Gc heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein | | Authors: | Du, S, Peng, R, Qi, J, Li, C. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.
Nat Commun, 14, 2023
|
|
8I4T
 
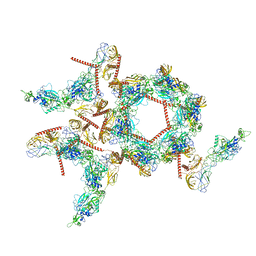 | | Structure of the asymmetric unit of SFTSV virion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein | | Authors: | Du, S, Peng, R, Qi, J, Li, C. | | Deposit date: | 2023-01-21 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.
Nat Commun, 14, 2023
|
|
8CY8
 
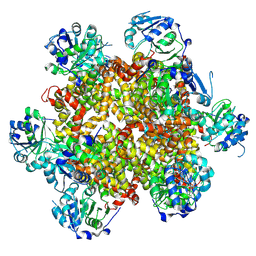 | |
5GGZ
 
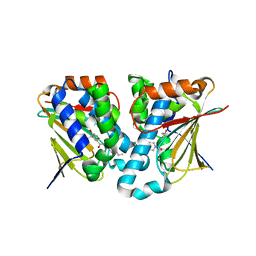 | | Crystal structure of novel inhibitor bound with Hsp90 | | Descriptor: | Heat shock protein HSP 90-alpha, [2,4-bis(oxidanyl)-5-propan-2-yl-phenyl]-(2-ethoxy-7,8-dihydro-5~{H}-pyrido[4,3-d]pyrimidin-6-yl)methanone | | Authors: | Chen, T.T, Li, J, Xu, Y.C. | | Deposit date: | 2016-06-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Novel Tetrahydropyrido[4,3-d]pyrimidines as Potent Inhibitors of Chaperone Heat Shock Protein 90
J. Med. Chem., 59, 2016
|
|
3FVH
 
 | |
3HIH
 
 | |
3HIK
 
 | | Structure of human Plk1-PBD in complex with PLHSpT | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pentamer phosphopeptide, ... | | Authors: | Wlodawer, A, Moulaei, T. | | Deposit date: | 2009-05-20 | | Release date: | 2009-06-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and functional analyses of minimal phosphopeptides targeting the polo-box domain of polo-like kinase 1.
Nat.Struct.Mol.Biol., 16, 2009
|
|
8AYH
 
 | |
3B7V
 
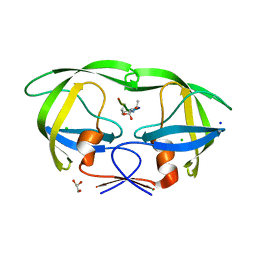 | | HIV-1 protease complexed with gem-diol-amine tetrahedral intermediate NLLTQI | | Descriptor: | CHLORIDE ION, GLYCEROL, Protease, ... | | Authors: | Kovalevsky, A.Y, Chumanevich, A.A, Weber, I.T. | | Deposit date: | 2007-10-31 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Caught in the Act: The 1.5 A Resolution Crystal Structures of the HIV-1 Protease and the I54V Mutant Reveal a Tetrahedral Reaction Intermediate.
Biochemistry, 46, 2007
|
|
3B80
 
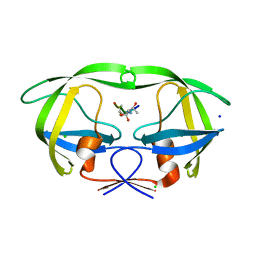 | | HIV-1 protease mutant I54V complexed with gem-diol-amine intermediate NLLTQI | | Descriptor: | CHLORIDE ION, Protease, SODIUM ION, ... | | Authors: | Chumanevich, A.A, Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2007-10-31 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Caught in the Act: The 1.5 A Resolution Crystal Structures of the HIV-1 Protease and the I54V Mutant Reveal a Tetrahedral Reaction Intermediate.
Biochemistry, 46, 2007
|
|
7C2K
 
 | | COVID-19 RNA-dependent RNA polymerase pre-translocated catalytic complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (29-MER), ... | | Authors: | Wang, Q, Gao, Y, Ji, W, Mu, A, Rao, Z. | | Deposit date: | 2020-05-07 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase.
Cell, 182, 2020
|
|
6A52
 
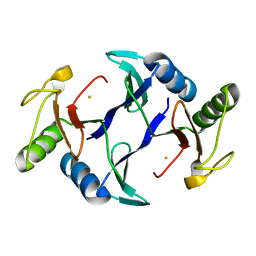 | | Oxidase ChaP-H1 | | Descriptor: | FE (II) ION, dioxidase ChaP-H1 | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
6LDZ
 
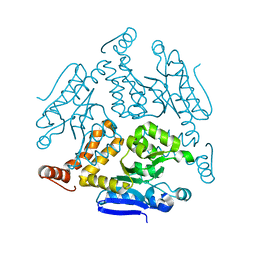 | | Crystal structure of Rv0222 from Mycobacterium tuberculosis | | Descriptor: | Probable enoyl-CoA hydratase EchA1 (Enoyl hydrase) (Unsaturated acyl-CoA hydratase) (Crotonase) | | Authors: | Li, J, Ran, Y.J, Wang, L, Wu, J.H, Ge, B.X, Rao, Z.H. | | Deposit date: | 2019-11-23 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Host-mediated ubiquitination of a mycobacterial protein suppresses immunity.
Nature, 577, 2020
|
|
6A4X
 
 | | Oxidase ChaP-H2 | | Descriptor: | Bleomycin resistance protein, FE (II) ION | | Authors: | Zhang, B, Wang, Y.S, Ge, H.M. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
6A4Z
 
 | | Oxidase ChaP | | Descriptor: | ChaP protein, FE (II) ION | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
