1ZKI
 
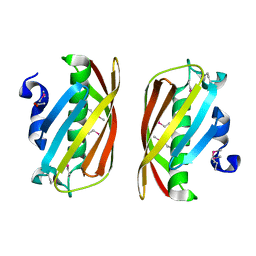 | | Structure of conserved protein PA5202 from Pseudomonas aeruginosa | | Descriptor: | ACETIC ACID, hypothetical protein PA5202 | | Authors: | Cuff, M.E, Evdokimova, E, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and activity of the Pseudomonas aeruginosa hotdog-fold thioesterases PA5202 and PA2801.
Biochem.J., 444, 2012
|
|
3SVI
 
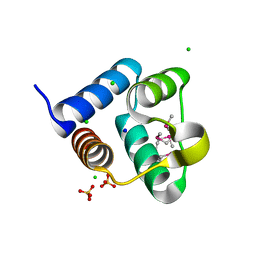 | | Structure of the Pto-binding domain of HopPmaL generated by limited thermolysin digestion | | Descriptor: | CHLORIDE ION, SODIUM ION, SULFATE ION, ... | | Authors: | Singer, A.U, Stein, A, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-07-12 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
2M2J
 
 | | Solution NMR structure of the N-terminal domain of STM1478 from Salmonella typhimurium LT2: Target STR147A of the Northeast Structural Genomics consortium (NESG), and APC101565 of the Midwest Center for Structural Genomics (MCSG). | | Descriptor: | Putative periplasmic protein | | Authors: | Houliston, S, Yee, A, Lemak, A, Garcia, M, Wu, B, Savchenko, A, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-21 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of DUF1471 Domains of Salmonella Proteins SrfN, YdgH/SssB, and YahO.
Plos One, 9, 2014
|
|
4WZ2
 
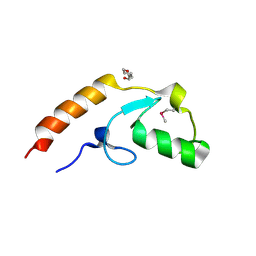 | | Crystal structure of U-box 2 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris, Ile175Met mutant | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase LubX, HEXANE-1,6-DIOL | | Authors: | Stogios, P.J, Qualie, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
3M16
 
 | | Structure of a Transaldolase from Oleispira antarctica | | Descriptor: | Transaldolase | | Authors: | Singer, A.U, Kagan, O, Zhang, R, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-04 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3EXC
 
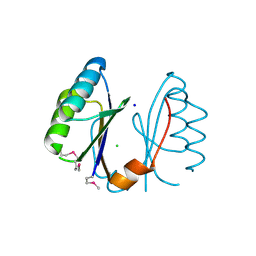 | | Structure of the RNA'se SSO8090 from Sulfolobus solfataricus | | Descriptor: | CHLORIDE ION, SODIUM ION, Uncharacterized protein | | Authors: | Singer, A.U, Skarina, T, Tan, K, Kagan, O, Onopriyenko, O, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the RNA'se SSO8090 from Sulfolobus solfataricus
To be Published
|
|
3EXM
 
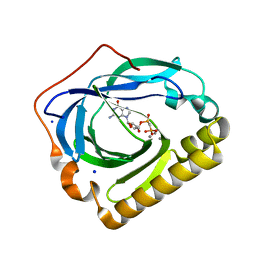 | | Crystal structure of the phosphatase SC4828 with the non-hydrolyzable nucleotide GPCP | | Descriptor: | CALCIUM ION, GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID GUANOSYL ESTER, ... | | Authors: | Singer, A.U, Xu, X, Zheng, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-16 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and mechanism of a new family of prokaryotic nucleoside diphosphatases.
To be Published
|
|
1NXH
 
 | | X-RAY STRUCTURE: NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT87 | | Descriptor: | MTH396 protein | | Authors: | Khayat, R, Savchenko, A, Edwards, A, Arowsmith, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-10 | | Release date: | 2004-02-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-RAY STRUCTURE OF MTH396
To be Published
|
|
3FIW
 
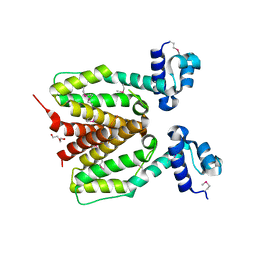 | | Structure of SCO0253, a Tetr-family transcriptional regulator from Streptomyces coelicolor | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative tetR-family transcriptional regulator | | Authors: | Singer, A.U, Xu, X, Chang, C, Gu, J, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-12 | | Release date: | 2009-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SCO0253, a Tetr-family transcriptional regulator from Streptomyces coelicolor
To be Published
|
|
4WZ3
 
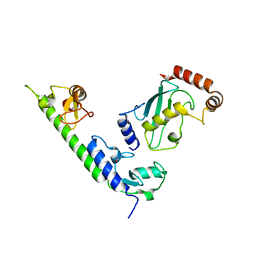 | | Crystal structure of the complex between LubX/LegU2/Lpp2887 U-box 1 and Homo sapiens UBE2D2 | | Descriptor: | E3 ubiquitin-protein ligase LubX, Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Stogios, P.J, Quaile, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-18 | | Release date: | 2015-01-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
7TQ1
 
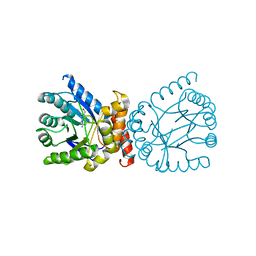 | | Crystal structure of adaptive laboratory evolved sulfonamide-resistant Dihydropteroate Synthase (DHPS) from Escherichia coli in complex with 6-hydroxymethylpterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, Dihydropteroate synthase | | Authors: | Stogios, P.J, Skarina, T, Tan, K, Venkatesan, M, Fruci, M, Joachimiak, A, Savchenko, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-26 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
1NN4
 
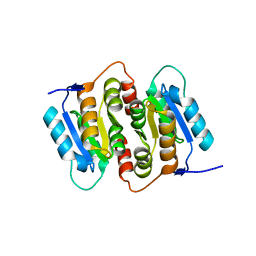 | | Structural Genomics, RpiB/AlsB | | Descriptor: | Ribose 5-phosphate isomerase B | | Authors: | Zhang, R.G, Andersson, C.E, Mowbray, S.L, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S.L, Arrowsmith, C, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-12 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A resolution structure of RpiB/AlsB from Escherichia coli illustrates a new approach to the ribose-5-phosphate isomerase reaction.
J.Mol.Biol., 332, 2003
|
|
3LMB
 
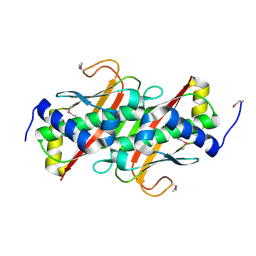 | | The crystal structure of the protein OLEI01261 with unknown function from Chlorobaculum tepidum TLS | | Descriptor: | Uncharacterized protein | | Authors: | Zhang, R, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3LNP
 
 | | Crystal Structure of Amidohydrolase family Protein OLEI01672_1_465 from Oleispira antarctica | | Descriptor: | ACETIC ACID, Amidohydrolase family Protein OLEI01672_1_465, CALCIUM ION, ... | | Authors: | Kim, Y, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3N3T
 
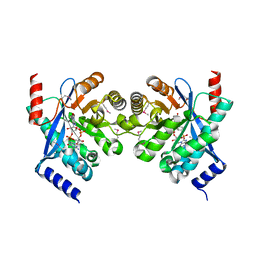 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase complex with cyclic di-gmp | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases.
J.Mol.Biol., 402, 2010
|
|
7TBU
 
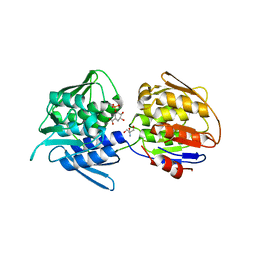 | | Crystal structure of the 5-enolpyruvate-shikimate-3-phosphate synthase (EPSPS) domain of Aro1 from Candida albicans in complex with shikimate-3-phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-enolpyruvylshikimate-3-phosphate synthase, SHIKIMATE-3-PHOSPHATE | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
7TBV
 
 | | Crystal structure of the shikimate kinase + 3-dehydroquinate dehydratase + 3-dehydroshikimate dehydrogenase domains of Aro1 from Candida albicans | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
3F4A
 
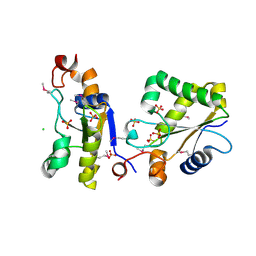 | | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family | | Descriptor: | AMMONIUM ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Osipiuk, J, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family
To be Published
|
|
5ES2
 
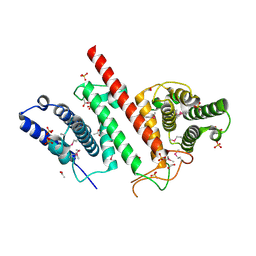 | | The crystal structure of a functionally uncharacterized protein LPG0634 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-16 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of a functionally uncharacterized protein LPG0634 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To Be Published
|
|
3FOV
 
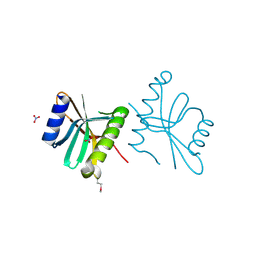 | | Crystal structure of protein RPA0323 of unknown function from Rhodopseudomonas palustris | | Descriptor: | NITRATE ION, UPF0102 protein RPA0323 | | Authors: | Osipiuk, J, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-02 | | Release date: | 2009-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | X-ray crystal structure of protein RPA0323 of unknown function from Rhodopseudomonas palustris.
To be Published
|
|
6V98
 
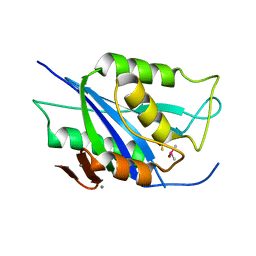 | | Crystal structure of Type VI secretion system effector, TseH (VCA0285) | | Descriptor: | CALCIUM ION, Cysteine hydrolase | | Authors: | Watanabe, N, Hersch, S.J, Dong, T.G, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Envelope stress responses defend against type six secretion system attacks independently of immunity proteins.
Nat Microbiol, 5, 2020
|
|
1O8B
 
 | | Structure of Escherichia coli ribose-5-phosphate isomerase, RpiA, complexed with arabinose-5-phosphate. | | Descriptor: | 5-O-phosphono-beta-D-arabinofuranose, RIBOSE 5-PHOSPHATE ISOMERASE | | Authors: | Zhang, R.-g, Andersson, C.E, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Mowbray, S.L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-11-26 | | Release date: | 2003-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of Escherichia Coli Ribose-5-Phosphate Isomerase: A Ubiquitous Enzyme of the Pentose Phosphate Pathway and the Calvin Cycle
Structure, 11, 2003
|
|
3F4F
 
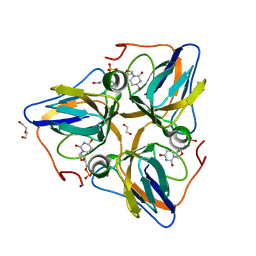 | | Crystal structure of dUT1p, a dUTPase from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
5DPO
 
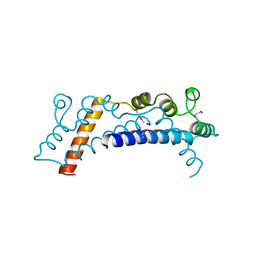 | |
3TJY
 
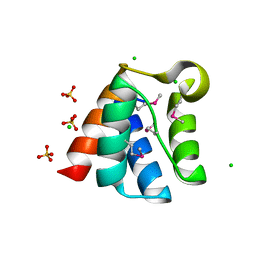 | | Structure of the Pto-binding domain of HopPmaL generated by limited chymotrypsin digestion | | Descriptor: | CHLORIDE ION, Effector protein hopAB3, SULFATE ION | | Authors: | Singer, A.U, Stein, A, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-08-25 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of HopPmaL reveals the presence of a second adaptor domain common to the HopAB family of Pseudomonas syringae type III effectors.
Biochemistry, 51, 2012
|
|
