2OPY
 
 | | Smac mimic bound to BIR3-XIAP | | Descriptor: | 1-({2-[(1S)-1-AMINOETHYL]-1,3-OXAZOL-4-YL}CARBONYL)-L-PROLYL-L-TRYPTOPHAN, Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Wist, A.D, Lu, G, Riedl, S.J, Shi, Y, McLendon, G.L. | | Deposit date: | 2007-01-30 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-activity based study of the Smac-binding pocket within the BIR3 domain of XIAP.
Bioorg.Med.Chem., 15, 2007
|
|
3ROO
 
 | | Murine class I major histocompatibility complex H-2Kb in complex with immunodominant LCMV-derived gp34-41 peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Madhurantakam, C, Duru, A.D, Leong, C, Sandalova, T, Webb, J.R, Achour, A. | | Deposit date: | 2011-04-26 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nitro-tyrosination of the immunodominant LCMV epitope gp34-41 alters both its capacity to stabilize H-2Kb and the molecular surface of the MHC complex, affecting TCR recognition
PLoS ONE, 2012
|
|
2OPZ
 
 | | AVPF bound to BIR3-XIAP | | Descriptor: | AVPF (Smac homologue, N-terminal tetrapeptide), Baculoviral IAP repeat-containing protein 4, ... | | Authors: | Wist, A.D. | | Deposit date: | 2007-01-30 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-activity based study of the Smac-binding pocket within the BIR3 domain of XIAP.
Bioorg.Med.Chem., 15, 2007
|
|
3OX2
 
 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | 2-hydroxy-8,9-dimethoxy-6H-isoindolo[2,1-a]indol-6-one, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Pegan, S.D, Sturdy, M, Mesecar, A.D. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
2P4V
 
 | | Crystal structure of the transcript cleavage factor, GreB at 2.6A resolution | | Descriptor: | Transcription elongation factor greB | | Authors: | Vassylyeva, M.N, Svetlov, V, Dearborn, A.D, Klyuyev, S, Artsimovitch, I, Vassylyev, D.G. | | Deposit date: | 2007-03-13 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The carboxy-terminal coiled-coil of the RNA polymerase beta'-subunit is the main binding site for Gre factors.
Embo Rep., 8, 2007
|
|
2PE6
 
 | | Non-covalent complex between human SUMO-1 and human Ubc9 | | Descriptor: | SUMO-conjugating enzyme UBC9, Small ubiquitin-related modifier 1 | | Authors: | Capili, A.D, Lima, C.D. | | Deposit date: | 2007-04-02 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Analysis of a Complex between SUMO and Ubc9 Illustrates Features of a Conserved E2-Ubl Interaction.
J.Mol.Biol., 369, 2007
|
|
3S7F
 
 | |
2LS5
 
 | | Solution structure of a putative protein disulfide isomerase from Bacteroides thetaiotaomicron | | Descriptor: | Uncharacterized protein | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a putative protein disulfide isomerase from Bacteroides thetaiotaomicron
To be Published
|
|
3S26
 
 | | Crystal Structure of Murine Siderocalin (Lipocalin 2, 24p3) | | Descriptor: | Neutrophil gelatinase-associated lipocalin, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Correnti, C, Bandaranayake, A.D, Strong, R.K. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-28 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Daedalus: a robust, turnkey platform for rapid production of decigram quantities of active recombinant proteins in human cell lines using novel lentiviral vectors.
Nucleic Acids Res., 39, 2011
|
|
2M96
 
 | |
3S7D
 
 | |
3S7B
 
 | | Structural Basis of Substrate Methylation and Inhibition of SMYD2 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, N-cyclohexyl-N~3~-[2-(3,4-dichlorophenyl)ethyl]-N-(2-{[2-(5-hydroxy-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-8-yl)ethyl]amino}ethyl)-beta-alaninamide, N-lysine methyltransferase SMYD2, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2011-05-26 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of Substrate Methylation and Inhibition of SMYD2.
Structure, 19, 2011
|
|
2L8T
 
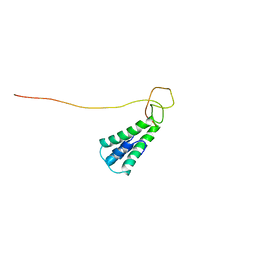 | | Staphylococcus aureus pathogenicity island 1 protein gp6, an internal scaffold in size determination | | Descriptor: | Transposon Tn557 toxic shock syndrome toxin-1 | | Authors: | Dearborn, A.D, Spilman, M.S, Damle, P.K, Chang, J.R, Monroe, E.B, Saad, J.S, Christie, G.E, Dokland, T. | | Deposit date: | 2011-01-24 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Staphylococcus aureus Pathogenicity Island 1 Protein gp6 Functions as an Internal Scaffold during Capsid Size Determination.
J.Mol.Biol., 412, 2011
|
|
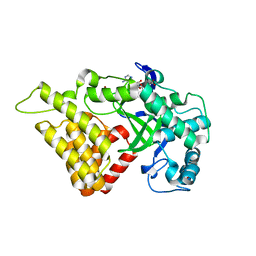
3S7J
 
 | |
2Q7M
 
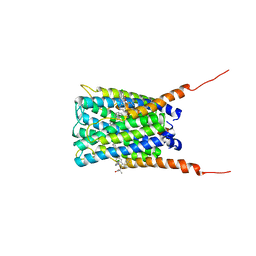 | | Crystal structure of human FLAP with MK-591 | | Descriptor: | 3-[3-(TERT-BUTYLTHIO)-1-(4-CHLOROBENZYL)-5-(QUINOLIN-2-YLMETHOXY)-1H-INDOL-2-YL]-2,2-DIMETHYLPROPANOIC ACID, Arachidonate 5-lipoxygenase-activating protein | | Authors: | Ferguson, A.D. | | Deposit date: | 2007-06-07 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein.
Science, 317, 2007
|
|
2Q7R
 
 | | Crystal structure of human FLAP with an iodinated analog of MK-591 | | Descriptor: | 3-[3-(3,3-DIMETHYLBUTANOYL)-1-(4-IODOBENZYL)-5-(QUINOLIN-2-YLMETHOXY)-1H-INDOL-2-YL]-2,2-DIMETHYLPROPANOIC ACID, Arachidonate 5-lipoxygenase-activating protein | | Authors: | Ferguson, A.D. | | Deposit date: | 2007-06-07 | | Release date: | 2007-08-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein.
Science, 317, 2007
|
|
2QX4
 
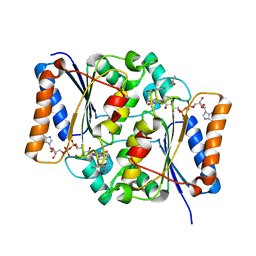 | | Crystal Structure of Quinone Reductase II | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Calamini, B, Santarsiero, B.D, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2007-08-10 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Biochem.J., 413, 2008
|
|
2QTN
 
 | | Crystal Structure of Nicotinate Mononucleotide Adenylyltransferase | | Descriptor: | GLYCEROL, MAGNESIUM ION, NICOTINATE MONONUCLEOTIDE, ... | | Authors: | Sershon, V.C, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2007-08-02 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and X-ray structural evidence for negative cooperativity in substrate binding to nicotinate mononucleotide adenylyltransferase (NMAT) from Bacillus anthracis.
J.Mol.Biol., 385, 2009
|
|
2QX8
 
 | | Crystal Structure of Quinone Reductase II | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Calamini, B, Santarsiero, B.D, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2007-08-10 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Biochem.J., 413, 2008
|
|
4QXI
 
 | | Crystal structure of human AR complexed with NADP+ and AK198 | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {2-[(4-amino-2-fluorobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Fanfrlik, J, Hobza, P, Podjarny, A.D. | | Deposit date: | 2014-07-21 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.867 Å) | | Cite: | The Effect of Halogen-to-Hydrogen Bond Substitution on Human Aldose Reductase Inhibition.
Acs Chem.Biol., 10, 2015
|
|
3MKJ
 
 | | Methionine gamma-lyase from Citrobacter freundii with pyridoximine-5'-phosphate | | Descriptor: | Methionine gamma-lyase, [5-hydroxy-4-(iminomethyl)-6-methyl-pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Revtovich, S.V, Nikulin, A.D, Morozova, E.A, Demidkina, T.V. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Exploring methionine gamma-lyase structure-function relationship via microspectrophotometry and X-ray crystallography
Biochim.Biophys.Acta, 1814, 2011
|
|
4RJL
 
 | | Gamma subunit of the translation initiation factor 2 from Sulfolobus solfataricus complexed with GDPCP | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Kravchenko, O.V, Nikonov, O.S, Arhipova, V.I, Stolboushkina, E.A, Gabdulkhakov, A.G, Nikulin, A.D, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2014-10-09 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6401 Å) | | Cite: | Crystal structure of gamma subunit of the translation initiation factor 2 from Sulfolobus solfataricus in complex with GDPCP at 1.64A resolution
To be Published
|
|
4RSP
 
 | |
3MB2
 
 | | Kinetic and Structural Characterization of a Heterohexamer 4-Oxalocrotonate Tautomerase from Chloroflexus aurantiacus J-10-fl: Implications for Functional and Structural Diversity in the Tautomerase Superfamily | | Descriptor: | 4-oxalocrotonate tautomerase family enzyme - alpha subunit, 4-oxalocrotonate tautomerase family enzyme - beta subunit, SULFATE ION | | Authors: | Burks, E.A, Fleming, C.D, Mesecar, A.D, Whitman, C.P, Pegan, S.D. | | Deposit date: | 2010-03-24 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Kinetic and structural characterization of a heterohexamer 4-oxalocrotonate tautomerase from Chloroflexus aurantiacus J-10-fl: implications for functional and structural diversity in the tautomerase superfamily
Biochemistry, 49, 2010
|
|
2V7A
 
 | | Crystal structure of the T315I Abl mutant in complex with the inhibitor PHA-739358 | | Descriptor: | MAGNESIUM ION, N-[(3E)-5-[(2R)-2-METHOXY-2-PHENYLACETYL]PYRROLO[3,4-C]PYRAZOL-3(5H)-YLIDENE]-4-(4-METHYLPIPERAZIN-1-YL)BENZAMIDE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ABL1 | | Authors: | Modugno, M, Casale, E, Soncini, C, Rosettani, P, Colombo, R, Lupi, R, Rusconi, L, Fancelli, D, Carpinelli, P, Cameron, A.D, Isacchi, A, Moll, J. | | Deposit date: | 2007-07-27 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the T315I Abl Mutant in Complex with the Aurora Kinases Inhibitor Pha-739358.
Cancer Res., 67, 2007
|
|
