6PZE
 
 | |
6PZF
 
 | |
2QGB
 
 | | Human transthyretin (TTR) in Apo-form | | Descriptor: | Transthyretin | | Authors: | Connelly, S, Wilson, I.A. | | Deposit date: | 2007-06-28 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and structural evaluation of highly selective 2-arylbenzoxazole-based transthyretin amyloidogenesis inhibitors.
J.Med.Chem., 51, 2008
|
|
6Q23
 
 | |
6PZG
 
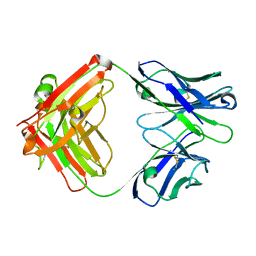 | | Crystal structure of human NA-80 Fab | | Descriptor: | NA-80 Fab heavy chain, NA-80 Fab light chain | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2019-07-31 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
2QGD
 
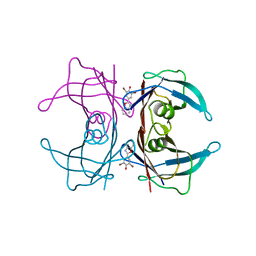 | |
2NTF
 
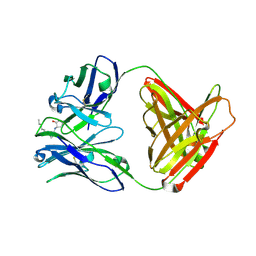 | |
6PZH
 
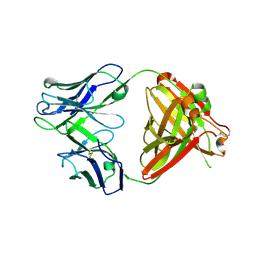 | | Crystal structure of human NA-22 Fab | | Descriptor: | NA-22 Fab heavy chain, NA-22 Fab light chain | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2019-07-31 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
2QSC
 
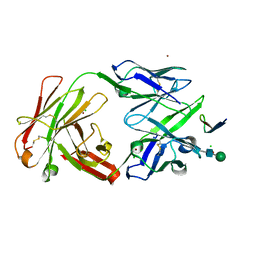 | | Crystal structure analysis of anti-HIV-1 V3-Fab F425-B4e8 in complex with a V3-peptide | | Descriptor: | CHLORIDE ION, Envelope glycoprotein gp120, Fab F425-B4e8, ... | | Authors: | Bell, C.H, Schiefner, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2007-07-30 | | Release date: | 2008-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of antibody F425-B4e8 in complex with a V3 peptide reveals a new binding mode for HIV-1 neutralization.
J.Mol.Biol., 375, 2008
|
|
6Q20
 
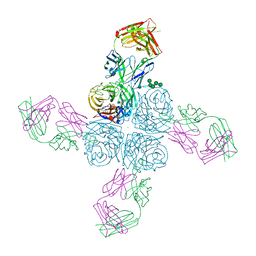 | |
2QGC
 
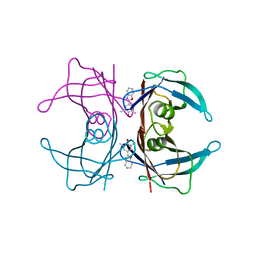 | |
2OJZ
 
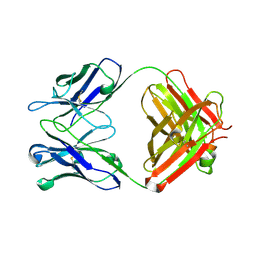 | | Anti-DNA antibody ED10 | | Descriptor: | Fab ED10 heavy chain, Fab ED10 light chain | | Authors: | Stanfield, R.L, Sanguineti, S, Wilson, I.A, de Prat-Gay, G. | | Deposit date: | 2007-01-15 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Specific recognition of a DNA immunogen by its elicited antibody
J.Mol.Biol., 370, 2007
|
|
2O5X
 
 | |
2OK0
 
 | | Fab ED10-DNA complex | | Descriptor: | 5'-D(*TP*C)-3', Fab ED10 heavy chain, Fab ED10 light chain | | Authors: | Stanfield, R.L, Sanguineti, S, Wilson, I.A, de Prat-Gay, G. | | Deposit date: | 2007-01-15 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Specific recognition of a DNA immunogen by its elicited antibody
J.Mol.Biol., 370, 2007
|
|
2O5Z
 
 | |
2OP4
 
 | | Crystal Structure of Quorum-Quenching Antibody 1G9 | | Descriptor: | 1,2-ETHANEDIOL, Murine Antibody Fab RS2-1G9 IGG1 Heavy Chain, Murine Antibody Fab RS2-1G9 Lambda Light Chain | | Authors: | Kirchdoerfer, R.N, Debler, E.W, Wilson, I.A. | | Deposit date: | 2007-01-26 | | Release date: | 2007-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structures of a Quorum-quenching Antibody.
J.Mol.Biol., 368, 2007
|
|
6OC7
 
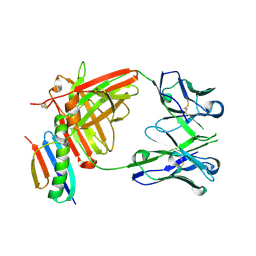 | | HMP42 Fab in complex with Protein G | | Descriptor: | Heavy chain of HMP42 Fab, Immunoglobulin G-binding protein G, Light chain for HMP42 Fab | | Authors: | Bernard, S.M, Wilson, I.A. | | Deposit date: | 2019-03-22 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses.
Science, 366, 2019
|
|
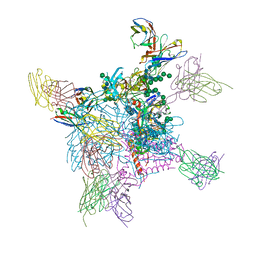
6OPA
 
 | |
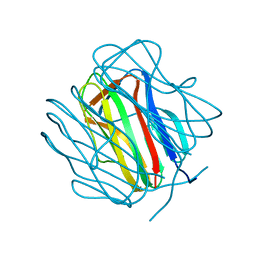
2R6Q
 
 | |
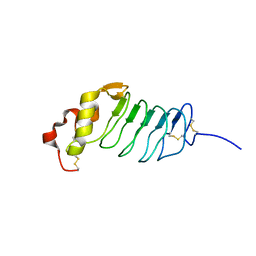
2R9U
 
 | |
2Q7Y
 
 | | Structure of the endogenous iNKT cell ligand iGb3 bound to mCD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Zajonc, D.M, Wilson, I.A, Teyton, L. | | Deposit date: | 2007-06-07 | | Release date: | 2008-04-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Mouse CD1d-iGb3 Complex and its Cognate Valpha14 T Cell Receptor Suggest a Model for Dual Recognition of Foreign and Self Glycolipids.
J.Mol.Biol., 377, 2008
|
|
2QGE
 
 | |
4M6L
 
 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADP+ and 5,10-dideazatetrahydrofolic acid | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Dihydrofolate reductase, N-(4-{2-[(6S)-2-amino-4-oxo-1,4,5,6,7,8-hexahydropyrido[2,3-d]pyrimidin-6-yl]ethyl}benzoyl)-L-glutamic acid, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4M5Y
 
 | | Crystal structure of broadly neutralizing Fab 5J8 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fab 5J8 heavy chain, Fab 5J8 light chain, ... | | Authors: | Lee, P.S, Wilson, I.A. | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Antibody Recognition of the Pandemic H1N1 Influenza Virus Hemagglutinin Receptor Binding Site.
J.Virol., 87, 2013
|
|
4MHH
 
 | |
