7UPX
 
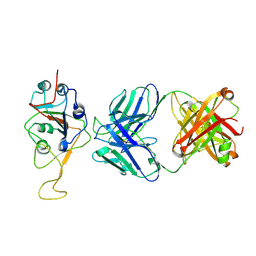 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, SP1-77 Fab heavy chain, SP1-77 Fab light chain, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
6LMS
 
 | |
7UPW
 
 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
2RBE
 
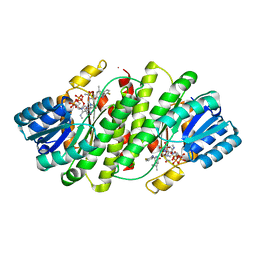 | | The discovery of 2-anilinothiazolones as 11beta-HSD1 inhibitors | | Descriptor: | (5R)-2-[(2-fluorophenyl)amino]-5-(1-methylethyl)-1,3-thiazol-4(5H)-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, J, Jordan, S.R, Li, V. | | Deposit date: | 2007-09-18 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The discovery of 2-anilinothiazolones as 11beta-HSD1 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4GVA
 
 | |
4GSB
 
 | |
5X5M
 
 | |
7X5V
 
 | |
9JA6
 
 | | Cryo-EM structure of Tdk1 tetramer complex | | Descriptor: | Meiotically up-regulated gene 135 protein,Immunoglobulin G-binding protein G | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2024-08-24 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural duality enables a single protein to act as a toxin-antidote pair for meiotic drive.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3SM4
 
 | |
3SLP
 
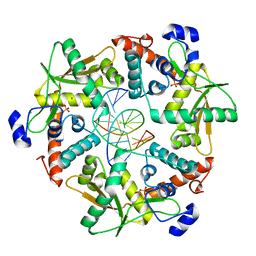 | |
8IW5
 
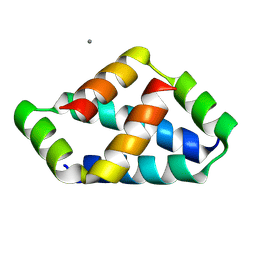 | | Crystal structure of liprin-beta H2H3 dimer | | Descriptor: | CALCIUM ION, Liprin-beta-1 | | Authors: | Zhang, J, Chen, S, Wei, Z. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
8IW0
 
 | | Crystal structure of the KANK1/liprin-beta1 complex | | Descriptor: | Liprin-beta-1,KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Zhang, J, Chen, S, Wei, Z, Yu, C. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
7DDE
 
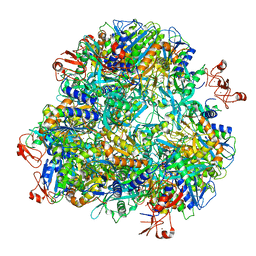 | | Cryo-EM structure of the Ape4 and Nbr1 complex | | Descriptor: | Aspartyl aminopeptidase 1,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
7DD9
 
 | | Cryo-EM structure of the Ams1 and Nbr1 complex | | Descriptor: | Alpha-mannosidase,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
7YFI
 
 | | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFG
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (major class in asymmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFH
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine, glutamate and (R)-PYD-106 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YUK
 
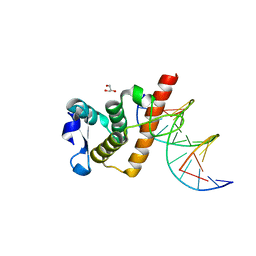 | | Complex structure of BANP BEN domain bound to DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*CP*GP*CP*GP*AP*GP*AP*G)-3'), GLYCEROL, Protein BANP | | Authors: | Zhang, J, Xiao, Y.Q, Chen, Y.X, Liu, K, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
7YUG
 
 | | Structure of human BANP BEN domain | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Zhang, J, Xiao, Y.Q, Chen, Y.X, Liu, K, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
7CWE
 
 | | Human Fructose-1,6-bisphosphatase 1 in APO R-state | | Descriptor: | Fructose-1,6-bisphosphatase 1, MAGNESIUM ION | | Authors: | Chen, Y, Zhang, J, Li, C, Cao, Y. | | Deposit date: | 2020-08-28 | | Release date: | 2021-09-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human Fructose-1,6-bisphosphatase 1 in APO R-state
To Be Published
|
|
6IMG
 
 | | Solution Structure of Bicyclic Peptide pb-13 | | Descriptor: | (ACE)-GLY-CYS-PRO-CYS-ILE-TRP-PRO-GLU-LEU-CYS-PRO-TRP-ILE-ARG-SER-CYS-(NH2) | | Authors: | Yao, H, Lin, P, Zha, J, Zha, M, Zhao, Y, Wu, C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Ordered and Isomerically Stable Bicyclic Peptide Scaffolds Constrained through Cystine Bridges and Proline Turns.
Chembiochem, 20, 2019
|
|
6IMH
 
 | | Solution Structure of Bicyclic Peptide pb-18 | | Descriptor: | (ACE)-GLY-CYS-PRO-CYS-GLU-PRO-SER-TYR-LEU-CYS-PRO-TRP-LEU-PRO-GLY-CYS-(NH2) | | Authors: | Yao, H, Lin, P, Zha, J, Zha, M, Zhao, Y, Wu, C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Ordered and Isomerically Stable Bicyclic Peptide Scaffolds Constrained through Cystine Bridges and Proline Turns.
Chembiochem, 20, 2019
|
|
4X2A
 
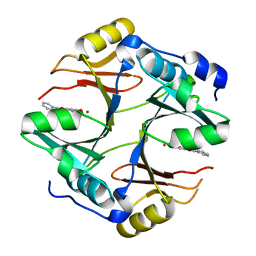 | | Crystal structure of mouse glyoxalase I complexed with baicalein | | Descriptor: | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhang, H, Zhai, J, Zhang, L, Li, C, Zhao, Y, Hu, X. | | Deposit date: | 2014-11-26 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In Vitro Inhibition of Glyoxalase І by Flavonoids: New Insights from Crystallographic Analysis.
Curr Top Med Chem, 16, 2016
|
|
4PV5
 
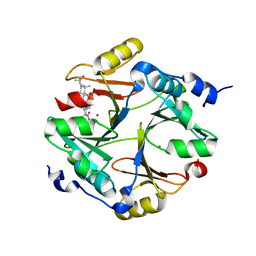 | | Crystal structure of mouse glyoxalase I in complexed with 18-beta-glycyrrhetinic acid | | Descriptor: | (3BETA,5BETA,14BETA)-3-HYDROXY-11-OXOOLEAN-12-EN-29-OIC ACID, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhang, H, Zhai, J, Zhang, L.P, Zhao, Y.N, Li, C, Hu, X.P. | | Deposit date: | 2014-03-15 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for 18-beta-glycyrrhetinic acid as a novel non-GSH analog glyoxalase I inhibitor
Acta Pharmacol.Sin., 36, 2015
|
|
