6JFX
 
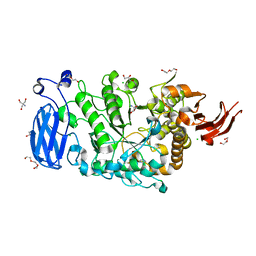 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with maltopentaose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-12 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6JHF
 
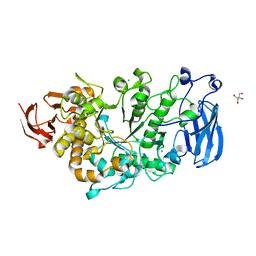 | | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6JFJ
 
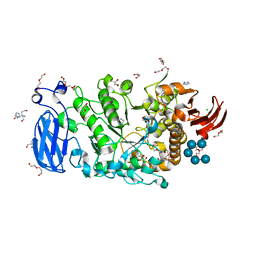 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with maltohexaose and alpha-cyclodextrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-09 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7D3I
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with MI-23 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[3-[3,5-bis(fluoranyl)phenyl]propanoyl]-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase | | Authors: | Zeng, R, Li, Y.S, Qiao, J.X, Wang, Y.F, Yang, S.Y, Lei, J. | | Deposit date: | 2020-09-19 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
6JEQ
 
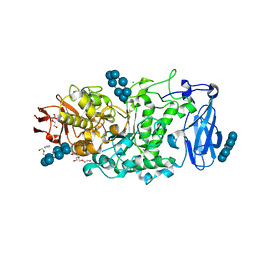 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with beta-cyclodextrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-07 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7COM
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Boceprevir (space group P212121) | | Descriptor: | 3C-like proteinase, boceprevir (bound form) | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Liu, J.M, Zhou, Y.L, Chen, P, Yang, S.Y, Lei, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7DFU
 
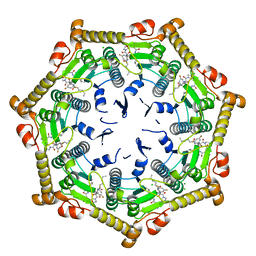 | |
7V6K
 
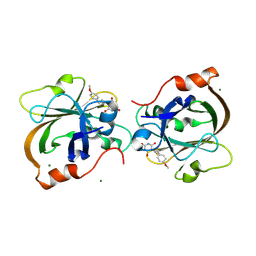 | | Crystal structure of Sortase A from Streptococcus pyogenes in complex with ML346 | | Descriptor: | 5-[3-(4-methoxyphenyl)prop-2-enylidene]-1,3-diazinane-2,4,6-trione, MAGNESIUM ION, Sortase | | Authors: | Yang, C.G, Gan, J. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Covalent sortase A inhibitor ML346 prevents Staphylococcus aureus infection of Galleria mellonella.
Rsc Med Chem, 13, 2022
|
|
5YUQ
 
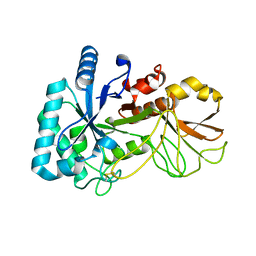 | | The high resolution structure of chitinase (RmChi1) from the thermophilic fungus Rhizomucor miehei (sp P1) | | Descriptor: | Chintase | | Authors: | Jiang, Z.Q, Hu, S.Q, Liu, Y.C, Qin, Z, Yan, Q.J, Yang, S.Q. | | Deposit date: | 2017-11-23 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Biochim Biophys Acta Proteins Proteom, 2021
|
|
7DFT
 
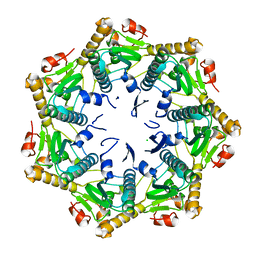 | | Crystal structure of Xanthomonas oryzae ClpP | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CHLORIDE ION | | Authors: | Yang, C.-G, Yang, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dysregulation of ClpP by Small-Molecule Activators Used Against Xanthomonas oryzae pv. oryzae Infections.
J.Agric.Food Chem., 69, 2021
|
|
2ZU3
 
 | | Complex structure of CVB3 3C protease with TG-0204998 | | Descriptor: | 3C proteinase, N-[(benzyloxy)carbonyl]-3-[(2,2-dimethylpropanoyl)amino]-L-alanyl-N-[(1R)-4-oxo-1-{[(3S)-2-oxopyrrolidin-3-yl]methyl}pentyl]-L-leucinamide | | Authors: | Lee, C.C, Tsui, Y.C, Wang, A.H.-J. | | Deposit date: | 2008-10-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
5YLH
 
 | | Structure of GH113 beta-1,4-mannanase | | Descriptor: | beta-1,4-mannanase | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase from Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
2ZU4
 
 | | Complex structure of SARS-CoV 3CL protease with TG-0204998 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-[(2,2-dimethylpropanoyl)amino]-L-alanyl-N-[(1R)-4-oxo-1-{[(3S)-2-oxopyrrolidin-3-yl]methyl}pentyl]-L-leucinamide | | Authors: | Hsu, M.F, Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2008-10-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
2ZTX
 
 | | Complex structure of CVB3 3C protease with EPDTC | | Descriptor: | 3C proteinase, zinc(II)hydrogensulfide | | Authors: | Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
2ZU5
 
 | | complex structure of SARS-CoV 3CL protease with TG-0205486 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-[(1R)-4-cyclopropyl-4-oxo-1-{[(3S)-2-oxopyrrolidin-3-yl]methyl}butyl]-L-leucinamide | | Authors: | Hsu, M.F, Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2008-10-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
2ZU1
 
 | |
2ZTY
 
 | |
8WTI
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with 20j | | Descriptor: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Zeng, R, Zhao, X, Yang, S.Y, Lei, J. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of alpha-Ketoamide inhibitors of SARS-CoV-2 main protease derived from quaternized P1 groups.
Bioorg.Chem., 143, 2024
|
|
2ZU2
 
 | | complex structure of CoV 229E 3CL protease with EPDTC | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3C-like proteinase, zinc(II)hydrogensulfide | | Authors: | Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2008-10-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
2ZTZ
 
 | |
5UBQ
 
 | | Cryo-EM structure of ciliary microtubule doublet | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ichikawa, M, Liu, D, Kastritis, P.L, Basu, K, Bui, K.H. | | Deposit date: | 2016-12-21 | | Release date: | 2017-05-10 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins.
Nat Commun, 8, 2017
|
|
5UCY
 
 | | Cryo-EM map of protofilament of microtubule doublet | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ichikawa, M, Liu, D, Kastritis, P.L, Basu, K, Bui, K.H. | | Deposit date: | 2016-12-22 | | Release date: | 2017-05-10 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins.
Nat Commun, 8, 2017
|
|
5U6K
 
 | |
8HUG
 
 | | F1 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 4-[4-(3-chlorophenyl)piperazin-1-yl]-3-[(3-fluorophenyl)sulfonylamino]benzoic acid, CHLORIDE ION, CRM1 isoform 1, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a Hidden Pocket beneath the NES Groove by Novel Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
8HUF
 
 | | B28 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 3-[(4-chlorophenyl)carbonylamino]-4-[4-(2,5-dimethylphenyl)piperazin-1-yl]benzoic acid, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of a Hidden Pocket beneath the NES Groove by Novel Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
