7SG6
 
 | | Structure of PfCSP peptide 21 with antibody CIS43_Var10 | | Descriptor: | CIS43_Var10 Fab Heavy chain, CIS43_Var10 Fab Light chain, GLYCEROL, ... | | Authors: | Tripathi, P, Kwong, P.D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Highly protective antimalarial antibodies via precision library generation and yeast display screening.
J.Exp.Med., 219, 2022
|
|
4RFN
 
 | | Crystal structure of ADCC-potent Rhesus macaque ANTIBODY JR4 in complex with HIV-1 CLADE A/E GP120 and M48 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB HEAVY CHAIN OF ADCC ANTI-HIV-1 ANTIBODY JR4, FAB LIGHT CHAIN OF ADCC ANTI-HIV-1 ANTIBODY JR4, ... | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2014-09-26 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
4R4B
 
 | | Crystal structure of the anti-hiv-1 antibody 2.2c | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 2.2C HEAVY CHAIN, FAB 2.2C LIGHT CHAIN, ... | | Authors: | McLellan, J.S, Acharya, P, Huang, C.-C, Robinson, J, Kwong, P.D. | | Deposit date: | 2014-08-19 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
4R4H
 
 | | Crystal structure of non-neutralizing, A32-like antibody 2.2c in complex with HIV-1 Env gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2.2c, Heavy chain, ... | | Authors: | Mclellan, J, Acharya, P, Huang, C.-C, Kwong, P.D. | | Deposit date: | 2014-08-19 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.28 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
4PSX
 
 | | Crystal structure of histone acetyltransferase complex | | Descriptor: | COENZYME A, Histone H3, Histone H4, ... | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2014-03-08 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
6M6W
 
 | | Crystal structure the toxin-antitoxin MntA-HpeT mutant-Y104A | | Descriptor: | Toxin-antitoxin system antidote Mnt family, Toxin-antitoxin system toxin HepN family | | Authors: | Ouyang, S.Y, Zhen, X.K. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Novel polyadenylylation-dependent neutralization mechanism of the HEPN/MNT toxin/antitoxin system.
Nucleic Acids Res., 48, 2020
|
|
4RFO
 
 | | Crystal structure of the ADCC-Potent Antibody N60-I3 Fab in complex with HIV-1 Clade A/E gp120 and M48u1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade A/E gp120, N60-i3 Fab heavy chain, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2014-09-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
6M6U
 
 | | Crystal structure the toxin-antitoxin MntA-HpeT mutant-D39ED41E | | Descriptor: | Toxin-antitoxin system antitoxin MntA family, Toxin-antitoxin system toxin HepN family | | Authors: | Ouyang, S.Y, Zhen, X.K. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Novel polyadenylylation-dependent neutralization mechanism of the HEPN/MNT toxin/antitoxin system.
Nucleic Acids Res., 48, 2020
|
|
4R4F
 
 | | Crystal structure of non-neutralizing, A32-like antibody 2.2c in complex with HIV-1 YU2 gp120 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2.2c LIGHT CHAIN, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2014-08-19 | | Release date: | 2014-09-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.514 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
6M6V
 
 | | Crystal structure the toxin-antitoxin MntA-HepT | | Descriptor: | RNA (5'-R(P*AP*AP*A)-3'), Toxin-antitoxin system antidote Mnt family, Toxin-antitoxin system toxin HepN family | | Authors: | Ouyang, S.Y, Zhen, X.K. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Novel polyadenylylation-dependent neutralization mechanism of the HEPN/MNT toxin/antitoxin system.
Nucleic Acids Res., 48, 2020
|
|
4R4N
 
 | | Crystal structure of the anti-hiv-1 antibody 2.2c in complex with hiv-1 93ug037 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2.2c LIGHT CHAIN, Antibody 2.2c heavy CHAIN, ... | | Authors: | Acharya, P, Louder, R, Kwong, P.D. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
4RFE
 
 | | Crystal structure of ADCC-potent ANTI-HIV-1 Rhesus macaque antibody JR4 Fab | | Descriptor: | CHLORIDE ION, Fab heavy chain of ADCC-potent anti-HIV-1 antibody JR4, Fab light chain of ADCC-potent anti-HIV-1 antibody JR4, ... | | Authors: | Wu, X, Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2014-09-25 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
7ENQ
 
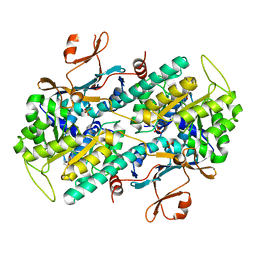 | | Crystal structure of human NAMPT in complex with compound NAT | | Descriptor: | 2-(2-~{tert}-butylphenoxy)-~{N}-(4-hydroxyphenyl)ethanamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Wang, G, Wu, C, Liu, M, Yao, H, Li, C, Wang, L, Tang, Y. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.204966 Å) | | Cite: | Discovery of small-molecule activators of nicotinamide phosphoribosyltransferase (NAMPT) and their preclinical neuroprotective activity.
Cell Res., 32, 2022
|
|
8HJE
 
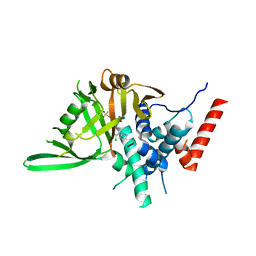 | | Vismodegib binds to the catalytical domain of human Ubiquitin-Specific Protease 28 | | Descriptor: | 2-chloranyl-~{N}-(4-chloranyl-3-pyridin-2-yl-phenyl)-4-methylsulfonyl-benzamide, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Shi, L, Wang, H, Xu, Z, Xiong, B, Zhang, N. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based discovery of potent USP28 inhibitors derived from Vismodegib.
Eur.J.Med.Chem., 254, 2023
|
|
9KET
 
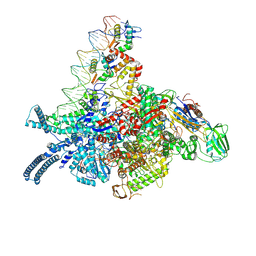 | |
9KEU
 
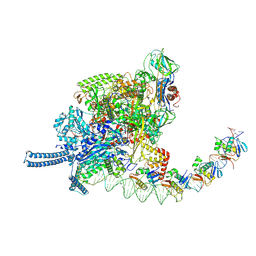 | |
9JI2
 
 | |
4CSG
 
 | |
4PTD
 
 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C MUTANT D274N | | Descriptor: | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis.
Biochemistry, 36, 1997
|
|
7CYB
 
 | | Saimiri boliviensis boliviensis galectin-13 with glycerol | | Descriptor: | GLYCEROL, Galectin | | Authors: | Su, J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Actin binding to galectin-13/placental protein-13 occurs independently of the galectin canonical ligand-binding site.
Glycobiology, 31, 2021
|
|
7CYA
 
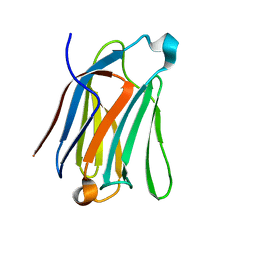 | | Saimiri boliviensis boliviensis galectin-13 with lactose | | Descriptor: | Galectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Actin binding to galectin-13/placental protein-13 occurs independently of the galectin canonical ligand-binding site.
Glycobiology, 31, 2021
|
|
7D14
 
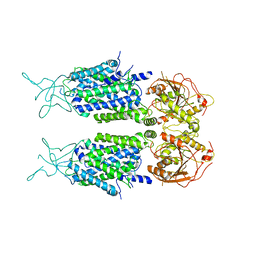 | | Mouse KCC2 | | Descriptor: | Solute carrier family 12 member 5 | | Authors: | Zhang, S, Yang, M. | | Deposit date: | 2020-09-13 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2.
Commun Biol, 4, 2021
|
|
7D10
 
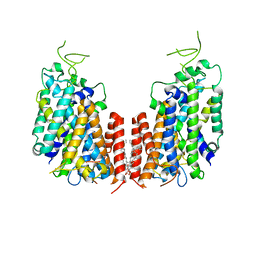 | | Human NKCC1 | | Descriptor: | PALMITIC ACID, Solute carrier family 12 member 2 | | Authors: | Zhang, S, Yang, M. | | Deposit date: | 2020-09-12 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2.
Commun Biol, 4, 2021
|
|
7FG9
 
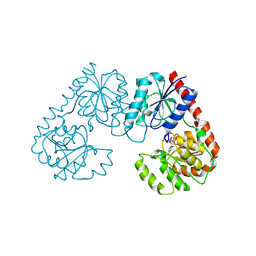 | | Alpha-1,2-glucosyltransferase_UDP_tll1591 | | Descriptor: | Glycosyl transferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Su, J.Y. | | Deposit date: | 2021-07-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for glucosylsucrose synthesis by a member of the alpha-1,2-glucosyltransferase family
Acta Biochim.Biophys.Sin., 54, 2022
|
|
7FGA
 
 | | Alpha-1,2-glucosyltransferase_UDP_sucrose_tll1591 | | Descriptor: | Glycosyl transferase, URIDINE-5'-DIPHOSPHATE, alpha-D-glucopyranose-(1-1)-alpha-D-fructofuranose | | Authors: | Su, J.Y. | | Deposit date: | 2021-07-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for glucosylsucrose synthesis by a member of the alpha-1,2-glucosyltransferase family
Acta Biochim.Biophys.Sin., 54, 2022
|
|
