3G7D
 
 | | Native PhpD with Cadmium Atoms | | Descriptor: | CADMIUM ION, PhpD | | Authors: | Nair, S.K. | | Deposit date: | 2009-02-09 | | Release date: | 2009-06-09 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An unusual carbon-carbon bond cleavage reaction during phosphinothricin biosynthesis.
Nature, 459, 2009
|
|
3LPI
 
 | | Structure of BACE Bound to SCH745132 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylsulfonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LPK
 
 | | Structure of BACE Bound to SCH747123 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N-[(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-{(2R)-4-[(3-methylphenyl)sulfonyl]piperazin-2-yl}ethyl]-3-{[(2R)-2-(methoxymethyl)pyrrolidin-1-yl]carbonyl}-5-methylbenzamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LNK
 
 | | Structure of BACE bound to SCH743813 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylcarbonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Orth, P, Cumming, J. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LPJ
 
 | | Structure of BACE Bound to SCH743641 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-[(1S,2S)-2-[(2R)-4-benzylpiperazin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7F6W
 
 | | Crystal structure of Saccharomyces cerevisiae lysyl-tRNA Synthetase | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, Lysine--tRNA ligase | | Authors: | Wu, S, Li, P, Hei, Z, Zheng, L, Wang, J, Fang, P. | | Deposit date: | 2021-06-26 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.607 Å) | | Cite: | Human lysyl-tRNA synthetase evolves a dynamic structure that can be stabilized by forming complex.
Cell.Mol.Life Sci., 79, 2022
|
|
5KVT
 
 | | THE STRUCTURE OF TRKA KINASE DOMAIN BOUND TO THE INHIBITOR ENTRECTINIB | | Descriptor: | Entrectinib, GLYCEROL, High affinity nerve growth factor receptor | | Authors: | Jin, L, Yan, S, Wei, G, Li, G, Harris, J, Vernier, J.-M. | | Deposit date: | 2016-07-15 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Antitumor Activity and Safety of the Pan-TRK, ROS1, and ALK inhibitor Entrectinib (RXDX-101): Combined Results from Two Phase I Trials
To Be Published
|
|
8K2P
 
 | | Crystal structure of CtGST-F76A | | Descriptor: | Glutathione S-transferase | | Authors: | Yang, J, Xiao, J.Y, Lei, X.G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
8K2O
 
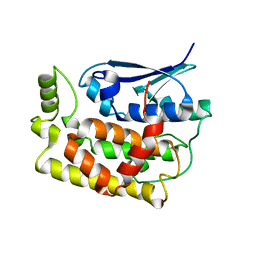 | | Crystal structure of Fhb7-M10 | | Descriptor: | Fhb7-M10 | | Authors: | Yang, J, Lei, X.G, Xiao, J.Y. | | Deposit date: | 2023-07-13 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
5CHL
 
 | | Structural basis of H2A.Z recognition by YL1 histone chaperone component of SRCAP/SWR1 chromatin remodeling complex | | Descriptor: | Histone H2A.Z, Vacuolar protein sorting-associated protein 72 homolog | | Authors: | Shan, S, Liang, X, Pan, L, Wu, C, Zhou, Z. | | Deposit date: | 2015-07-10 | | Release date: | 2016-03-09 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1
Nat.Struct.Mol.Biol., 23, 2016
|
|
3TG6
 
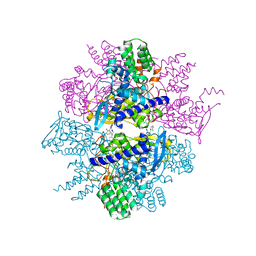 | | Crystal Structure of Influenza A Virus nucleoprotein with Ligand | | Descriptor: | Nucleocapsid protein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl][3-(2-chloropyridin-3-yl)-5-methyl-1,2-oxazol-4-yl]methanone | | Authors: | Pearce, B.C, Lewis, H.A, McDonnell, P.A, Steinbacher, S, Kiefersauer, R, Mortl, M, Maskos, K, Edavettal, S, Baldwin, E.T, Langley, D.R. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biophysical and Structural Characterization of a Novel Class of Influenza Virus Inhibitors
To be Published
|
|
3RO5
 
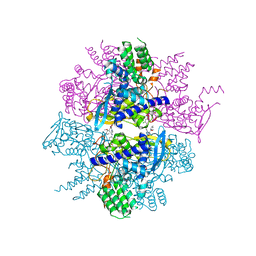 | | Crystal structure of influenza A virus nucleoprotein with ligand | | Descriptor: | Nucleocapsid protein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl][3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]methanone | | Authors: | Pearce, B.C, Edavettal, S, McDonnell, P.A, Lewis, H.A, Steinbacher, S, Baldwin, E.T, Langley, D.R, Maskos, K, Mortl, M, Kiefersauer, R. | | Deposit date: | 2011-04-25 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Inhibition of influenza virus replication via small molecules that induce the formation of higher-order nucleoprotein oligomers.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7KVZ
 
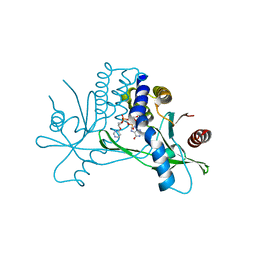 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN-2 | | Descriptor: | (2R,5R,7R,8R,10S,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2,10,16-trihydroxy-14-[(pyrimidin-4-yl)oxy]decahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R.J. | | Deposit date: | 2020-11-29 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
7KW1
 
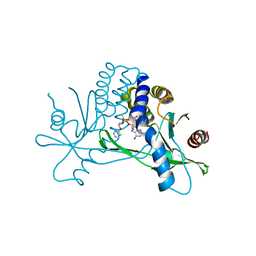 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN-3 | | Descriptor: | (2R,5R,7R,8R,10R,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-16-hydroxy-14-[(pyrimidin-4-yl)oxy]-2,10-disulfanyldecahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R. | | Deposit date: | 2020-11-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
7KVX
 
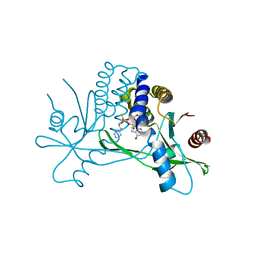 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN 1 | | Descriptor: | (2R,5R,7R,8R,10R,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-16-hydroxy-14-[(pyrimidin-4-yl)amino]-2,10-disulfanyldecahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R. | | Deposit date: | 2020-11-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
2L89
 
 | |
3RDT
 
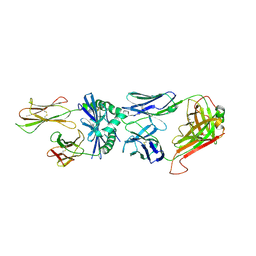 | | Crystal Structure of 809.B5 TCR complexed with MHC Class II I-Ab/3k peptide | | Descriptor: | 3K peptide, linker and MHC H-2 class II I-Ab beta chain, H-2 class II histocompatibility antigen, ... | | Authors: | Trenh, P, Huseby, E.S, Stern, L.J. | | Deposit date: | 2011-04-01 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A role for differential variable gene pairing in creating T cell receptors specific for unique major histocompatibility ligands.
Immunity, 35, 2011
|
|
4P5T
 
 | | 14.C6 TCR complexed with MHC class II I-Ab/3K peptide | | Descriptor: | H-2 class II histocompatibility antigen, A-B alpha chain, Human nkt tcr beta chain, ... | | Authors: | Trenh, P, Stadinski, B, Huseby, E.S, Stern, L.J. | | Deposit date: | 2014-03-19 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.263 Å) | | Cite: | Effect of CDR3 Sequences and Distal V Gene Residues in Regulating TCR-MHC Contacts and Ligand Specificity.
J Immunol., 192, 2014
|
|
5H49
 
 | | Crystal structure of Cbln1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
4P46
 
 | | J809.B5 Y31A TCR bound to IAb3K | | Descriptor: | 3K Peptide,H-2 class II histocompatibility antigen, A beta chain, H-2 class II histocompatibility antigen, ... | | Authors: | Stadinski, B.D, Huseby, E.S, Trenh, P, Stern, L.J. | | Deposit date: | 2014-03-11 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Effect of CDR3 Sequences and Distal V Gene Residues in Regulating TCR-MHC Contacts and Ligand Specificity.
J Immunol., 192, 2014
|
|
4P23
 
 | | J809.B5 TCR bound to IAb/3K | | Descriptor: | 3K peptide and MHC IAb beta chain,H-2 class II histocompatibility antigen, A beta chain, H-2 class II histocompatibility antigen, ... | | Authors: | Stadinski, B.D, Huseby, E.S, Trenh, P, Stern, L. | | Deposit date: | 2014-02-28 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Effect of CDR3 Sequences and Distal V Gene Residues in Regulating TCR-MHC Contacts and Ligand Specificity.
J Immunol., 192, 2014
|
|
5H48
 
 | | Crystal structure of Cbln1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5H4C
 
 | | Crystal structure of Cbln4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein Cbln4 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
7Y04
 
 | | Hsp90-AhR-p23 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aryl hydrocarbon receptor, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-06-03 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
7YOC
 
 | | Crystal structure of Fhb7 | | Descriptor: | Fhb7 | | Authors: | Yang, J, Liang, K, Xiao, J.Y, Lei, X.G. | | Deposit date: | 2022-08-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
