3D6H
 
 | |
3DGY
 
 | |
8RWK
 
 | | cryoEM structure of the central Ald4 filament determined by FilamentID | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium-activated aldehyde dehydrogenase, mitochondrial | | Authors: | Hugener, J, Xu, J, Wettstein, R, Ioannidi, L, Velikov, D, Wollweber, F, Henggeler, A, Matos, J, Pilhofer, M. | | Deposit date: | 2024-02-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | FilamentID reveals the composition and function of metabolic enzyme polymers during gametogenesis.
Cell, 187, 2024
|
|
2IEL
 
 | | CRYSTAL STRUCTURE OF TT0030 from Thermus Thermophilus | | Descriptor: | Hypothetical Protein TT0030 | | Authors: | Zhu, J, Huang, J, Stepanyuk, G, Chen, L, Chang, J, Zhao, M, Xu, H, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-09-19 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF TT0030 from Thermus Thermophilus AT 1.6 ANGSTROMS RESOLUTION
To be Published
|
|
3DH2
 
 | |
3D8A
 
 | | Co-crystal structure of TraM-TraD complex. | | Descriptor: | Protein traD, Relaxosome protein TraM | | Authors: | Glover, J.N.M, Lu, J, Wong, J.J, Edwards, R.A. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation.
Mol.Microbiol., 70, 2008
|
|
3S91
 
 | | Crystal Structure of the first bromodomain of human BRD3 in complex with the inhibitor JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Bromodomain-containing protein 3, ISOPROPYL ALCOHOL | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Keates, T, Felletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bradner, J.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD3 in complex with the inhibitor JQ1
To be Published
|
|
3S92
 
 | | Crystal Structure of the second bromodomain of human BRD3 in complex with the inhibitor JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, 1,2-ETHANEDIOL, Bromodomain-containing protein 3 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Keates, T, Felletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bradner, J.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal Structure of the second bromodomain of human BRD3 in complex with the inhibitor JQ1
To be Published
|
|
3SK0
 
 | | structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA mutant DhaA12 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Koudelakova, T, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2011-06-22 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dynamics and hydration explain failed functional transformation in dehalogenase design.
Nat.Chem.Biol., 10, 2014
|
|
2J91
 
 | | Crystal structure of Human Adenylosuccinate Lyase in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENYLOSUCCINATE LYASE, CHLORIDE ION, ... | | Authors: | Stenmark, P, Moche, M, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Johansson, I, Karlberg, T, Kosinska, U, Kotenyova, T, Magnusdottir, A, Nilsson, M.E, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Uppsten, M, Thorsell, A.G, van Den Berg, S, Wallden, K, Weigelt, J, Nordlund, P. | | Deposit date: | 2006-11-01 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Adenylosuccinate Lyase
To be Published
|
|
6Z97
 
 | | Structure of the prefusion SARS-CoV-2 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Duyvesteyn, H.M.E, Ren, J, Zhao, Y, Zhou, D, Huo, J, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-06-03 | | Release date: | 2020-07-01 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
5D7J
 
 | | Structure of human MR1-5-OP-RU in complex with human MAIT M33.64(Y95alphaF) TCR | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Keller, A.N, Woolley, R.E, Rossjohn, J. | | Deposit date: | 2015-08-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Diversity of T Cells Restricted by the MHC Class I-Related Molecule MR1 Facilitates Differential Antigen Recognition.
Immunity, 44, 2016
|
|
9QZT
 
 | |
3S93
 
 | | Crystal structure of conserved motif in TDRD5 | | Descriptor: | Tudor domain-containing protein 5, UNKNOWN ATOM OR ION | | Authors: | Chao, X, Tempel, W, Bian, C, Kania, J, Wernimont, A.K, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-31 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of conserved motif in TDRD5
to be published
|
|
5J9Y
 
 | | EGFR-T790M in complex with pyrazolopyrimidine inhibitor 1b | | Descriptor: | (R)-1-(3-(4-amino-3-(naphthalen-1-yl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl)piperidin-1-yl)prop-2-en-1-one, Epidermal growth factor receptor | | Authors: | Becker, C, Engel, J, Rauh, D. | | Deposit date: | 2016-04-11 | | Release date: | 2016-08-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insight into the Inhibition of Drug-Resistant Mutants of the Receptor Tyrosine Kinase EGFR.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
4LMR
 
 | | Crystal structure of L-lactate dehydrogenase from Bacillus cereus ATCC 14579, NYSGRC Target 029452 | | Descriptor: | L-lactate dehydrogenase 1 | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-10 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of L-lactate dehydrogenase from Bacillus cereus ATCC 14579, NYSGRC Target 029452
To be Published
|
|
6YQ4
 
 | | Crystal structure of Fusobacterium nucleatum tannase | | Descriptor: | GLYCEROL, MAGNESIUM ION, SPERMIDINE, ... | | Authors: | Mancheno, J.M, Anguita, J, Rodriguez, H. | | Deposit date: | 2020-04-16 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | A structurally unique Fusobacterium nucleatum tannase provides detoxicant activity against gallotannins and pathogen resistance.
Microb Biotechnol, 15, 2022
|
|
6Y4M
 
 | | Structure of Tubulin Tyrosine Ligase in Complex with Tb111 | | Descriptor: | (2~{R})-1-methylpiperidine-2-carboxylic acid, (2~{S},4~{R})-4-azanyl-2-methyl-5-phenyl-pentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Gavrilyuk, J, Nocek, B, Rigol, S, Nicolaou, K.C, Stoll, V. | | Deposit date: | 2020-02-21 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Tubulysin Analogues, Linker-Drugs, and Antibody-Drug Conjugates, Insights into Structure-Activity Relationships, and Tubulysin-Tubulin Binding Derived from X-ray Crystallographic Analysis.
J.Org.Chem., 86, 2021
|
|
4LBX
 
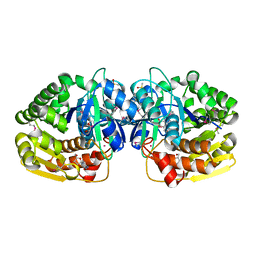 | | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, ADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium Etli CFN 42 complexed with cytidine
To be Published
|
|
6Y4N
 
 | | Structure of Tubulin Tyrosine Ligase in Complex with Tb116 | | Descriptor: | (2~{R})-1-methylpiperidine-2-carboxylic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Gavrilyuk, J, Nocek, B, Rigol, S, Nicolaou, K.C, Stoll, V. | | Deposit date: | 2020-02-21 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Tubulysin Analogues, Linker-Drugs, and Antibody-Drug Conjugates, Insights into Structure-Activity Relationships, and Tubulysin-Tubulin Binding Derived from X-ray Crystallographic Analysis.
J.Org.Chem., 86, 2021
|
|
3K71
 
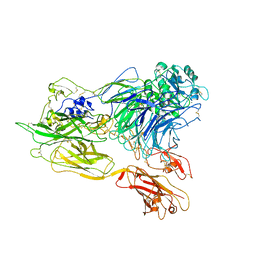 | | Structure of integrin alphaX beta2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xie, C, Zhu, J, Chen, X, Mi, L, Nishida, N, Springer, T.A. | | Deposit date: | 2009-10-11 | | Release date: | 2010-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of an integrin with an alphaI domain, complement receptor type 4.
Embo J., 29, 2010
|
|
5D39
 
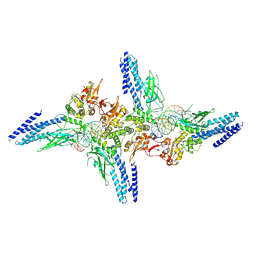 | | Transcription factor-DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*GP*GP*AP*AP*GP*AP*CP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*GP*TP*CP*TP*TP*CP*CP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-08-06 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D7L
 
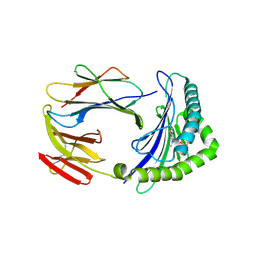 | | Structure of human MR1-5-OP-RU in complex with human MAV36 TCR | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, MAV36 TCR Alpha Chain, ... | | Authors: | Keller, A.N, Rossjohn, J. | | Deposit date: | 2015-08-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Diversity of T Cells Restricted by the MHC Class I-Related Molecule MR1 Facilitates Differential Antigen Recognition.
Immunity, 44, 2016
|
|
8S2X
 
 | |
8S2V
 
 | |
