6PWR
 
 | |
7YV7
 
 | |
7YV2
 
 | |
7F8L
 
 | |
7F5F
 
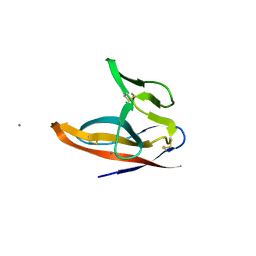 | | SARS-CoV-2 ORF8 S84 | | Descriptor: | CALCIUM ION, ORF8 protein | | Authors: | Chen, S, Zhou, Z, Chen, X. | | Deposit date: | 2021-06-22 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structures of Bat and Human Coronavirus ORF8 Protein Ig-Like Domain Provide Insights Into the Diversity of Immune Responses.
Front Immunol, 12, 2021
|
|
1J0Q
 
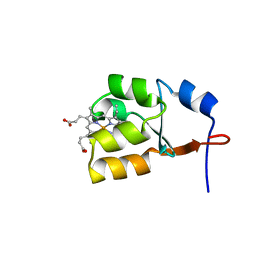 | | Solution Structure of Oxidized Bovine Microsomal Cytochrome b5 mutant V61H | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Wu, H, Huang, Z, Cao, C, Zhang, Q, Wang, Y.-H, Ma, J.-B, Xue, L.-L. | | Deposit date: | 2002-11-20 | | Release date: | 2003-08-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the oxidized bovine microsomal cytochrome b5 mutant V61H
Biochem.Biophys.Res.Commun., 307, 2003
|
|
8TS0
 
 | |
8URF
 
 | |
7E3O
 
 | |
6KLS
 
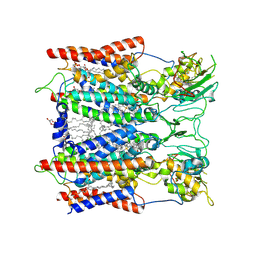 | | Hyperthermophilic respiratory Complex III | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, Cytochrome c, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guohong, P, Guoliang, Z, Hui, Z, Shuangbo, Z, Xiaoyun, P, Yan, Z. | | Deposit date: | 2019-07-30 | | Release date: | 2020-05-13 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A 3.3 angstrom -Resolution Structure of Hyperthermophilic Respiratory Complex III Reveals the Mechanism of Its Thermal Stability.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5UI0
 
 | |
5UHN
 
 | |
5UGX
 
 | |
5UGL
 
 | |
5JWC
 
 | | Structure of NDH2 from plasmodium falciparum in complex with RYL-552 | | Descriptor: | 5-fluoro-3-methyl-2-{4-[4-(trifluoromethoxy)benzyl]phenyl}quinolin-4(1H)-one, FLAVIN-ADENINE DINUCLEOTIDE, FRAGMENT OF TRITON X-100, ... | | Authors: | Yu, Y. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Target Elucidation by Cocrystal Structures of NADH-Ubiquinone Oxidoreductase of Plasmodium falciparum (PfNDH2) with Small Molecule To Eliminate Drug-Resistant Malaria
J. Med. Chem., 60, 2017
|
|
5JR9
 
 | | Crystal structure of apo-NeC3PO | | Descriptor: | NEQ131 | | Authors: | Zhang, J, Gan, J. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for single-stranded RNA recognition and cleavage by C3PO
Nucleic Acids Res., 44, 2016
|
|
5JWA
 
 | | the structure of malaria PfNDH2 | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, FRAGMENT OF TRITON X-100, ... | | Authors: | Yu, Y, Yang, Y.Q, Li, X.L, Yu, J, Ge, J.P, Li, J, Rao, Y, Yang, M.J. | | Deposit date: | 2016-05-11 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Target Elucidation by Cocrystal Structures of NADH-Ubiquinone Oxidoreductase of Plasmodium falciparum (PfNDH2) with Small Molecule To Eliminate Drug-Resistant Malaria
J. Med. Chem., 60, 2017
|
|
5JWB
 
 | | Structure of NDH2 from plasmodium falciparum in complex with NADH | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yu, Y, Li, X.L. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Target Elucidation by Cocrystal Structures of NADH-Ubiquinone Oxidoreductase of Plasmodium falciparum (PfNDH2) with Small Molecule To Eliminate Drug-Resistant Malaria
J. Med. Chem., 60, 2017
|
|
5JRC
 
 | |
5JRE
 
 | | Crystal structure of NeC3PO in complex with ssDNA. | | Descriptor: | 9-METHYL-9H-PURIN-6-AMINE, ADENINE, NEQ131, ... | | Authors: | Gan, J, Zhang, J. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for single-stranded RNA recognition and cleavage by C3PO
Nucleic Acids Res., 44, 2016
|
|
6UYA
 
 | | Crystal structure of Compound 19 bound to IRAK4 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{2-[(2R)-2-fluoro-3-hydroxy-3-methylbutyl]-6-(morpholin-4-yl)-1-oxo-2,3-dihydro-1H-isoindol-5-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide, SULFATE ION | | Authors: | Kiefer, J.R, Bryan, M.C, Lupardus, P.J, Zarrin, A.A, Rajapaksa, N.S, Gobbi, A, Drobnick, J, Kolesnikov, A, Liang, J, Do, S. | | Deposit date: | 2019-11-12 | | Release date: | 2019-11-20 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Potent Benzolactam IRAK4 Inhibitors with Robust in Vivo Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
6K1M
 
 | | Engineered form of a putative cystathionine gamma-lyase | | Descriptor: | Cystathionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE, PYRUVIC ACID | | Authors: | Chen, S, Wang, Y. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural characterization of cystathionine gamma-lyase smCSE enables aqueous metal quantum dot biosynthesis.
Int.J.Biol.Macromol., 174, 2021
|
|
6K1O
 
 | | Apo form of a putative cystathionine gamma-lyase | | Descriptor: | Cystathionine gamma-lyase | | Authors: | Chen, S, Wang, Y. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Structural characterization of cystathionine gamma-lyase smCSE enables aqueous metal quantum dot biosynthesis.
Int.J.Biol.Macromol., 174, 2021
|
|
6K1L
 
 | | E53A mutant of a putative cystathionine gamma-lyase | | Descriptor: | Cystathionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE, PYRUVIC ACID | | Authors: | Chen, S, Wang, Y. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural characterization of cystathionine gamma-lyase smCSE enables aqueous metal quantum dot biosynthesis.
Int.J.Biol.Macromol., 174, 2021
|
|
6K1N
 
 | | PLP-bound form of a putative cystathionine gamma-lyase | | Descriptor: | Cystathionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, S, Wang, Y. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural characterization of cystathionine gamma-lyase smCSE enables aqueous metal quantum dot biosynthesis.
Int.J.Biol.Macromol., 174, 2021
|
|
