1I6S
 
 | | T4 LYSOZYME MUTANT C54T/C97A/N101A | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Kovall, R.A, Baldwin, E.P, Matthews, B.W. | | Deposit date: | 2001-03-04 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
6A7F
 
 | |
5VVR
 
 | |
5VVS
 
 | | RNA pol II elongation complex | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Lahiri, I, Leschziner, A.E. | | Deposit date: | 2017-05-20 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Structural basis for the initiation of eukaryotic transcription-coupled DNA repair.
Nature, 551, 2017
|
|
7FEB
 
 | |
7FED
 
 | |
7FEC
 
 | |
2HHA
 
 | | The structure of DPP4 in complex with an oxadiazole inhibitor | | Descriptor: | (2S,3S)-3-{3-[4-(METHYLSULFONYL)PHENYL]-1,2,4-OXADIAZOL-5-YL}-1-OXO-1-PYRROLIDIN-1-YLBUTAN-2-AMINE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2006-06-28 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of potent, selective, and orally bioavailable oxadiazole-based dipeptidyl peptidase IV inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
7EJE
 
 | | human RAD51 post-synaptic complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Zhao, L.Y, Xu, J.F, Wang, H.W. | | Deposit date: | 2021-04-02 | | Release date: | 2022-03-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Mechanisms of distinctive mismatch tolerance between Rad51 and Dmc1 in homologous recombination.
Nucleic Acids Res., 49, 2021
|
|
7EJC
 
 | | human RAD51 presynaptic complex | | Descriptor: | 4-bromanyl-N-(4-bromophenyl)-3-[(phenylmethyl)sulfamoyl]benzamide, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Zhao, L.Y, Xu, J.F, Wang, H.W. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Mechanisms of distinctive mismatch tolerance between Rad51 and Dmc1 in homologous recombination.
Nucleic Acids Res., 49, 2021
|
|
7EJ6
 
 | | Yeast Dmc1 presynaptic complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), HLJ1_G0016300.mRNA.1.CDS.1, ... | | Authors: | Zhao, L.Y, Xu, J.F, Wang, H.W. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Mechanisms of distinctive mismatch tolerance between Rad51 and Dmc1 in homologous recombination.
Nucleic Acids Res., 49, 2021
|
|
7EJ7
 
 | | Yeast Dmc1 post-synaptic complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Zhao, L.Y, Xu, J.F, Wang, H.W. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Mechanisms of distinctive mismatch tolerance between Rad51 and Dmc1 in homologous recombination.
Nucleic Acids Res., 49, 2021
|
|
7XSV
 
 | | Crystal Structures of PIM1 in Complex with Macrocyclic Compound H3 | | Descriptor: | 8-Methyl-2,5,20-trioxa-8,13,17-triazatetracyclo[11.10.2.014,19.021,25]pentacosa-1(24),14(19),15,17,21(25),22-hexaene, Serine/threonine-protein kinase pim-1 | | Authors: | Shen, C, Xie, Y, Ren, X, Zhou, Y, Niu, H. | | Deposit date: | 2022-05-15 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Design, synthesis, and bioactivity evaluation of macrocyclic benzo[b]pyrido[4,3-e][1,4]oxazine derivatives as novel Pim-1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 72, 2022
|
|
3C43
 
 | | Human dipeptidyl peptidase IV/CD26 in complex with a flouroolefin inhibitor | | Descriptor: | (2S,3S)-4-cyclopropyl-3-{(3R,5R)-3-[2-fluoro-4-(methylsulfonyl)phenyl]-1,2,4-oxadiazolidin-5-yl}-1-[(3S)-3-fluoropyrrolidin-1-yl]-1-oxobutan-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Edmondson, S.D, Weber, A.E. | | Deposit date: | 2008-01-29 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fluoroolefins as amide bond mimics in dipeptidyl peptidase IV inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7YQE
 
 | |
7YSE
 
 | | Crystal structure of E. coli heterotetrameric GlyRS in complex with tRNA | | Descriptor: | Glycine--tRNA ligase alpha subunit, Glycine--tRNA ligase beta subunit, MAGNESIUM ION, ... | | Authors: | Han, L, Ju, Y, Zhou, H. | | Deposit date: | 2022-08-12 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | The binding mode of orphan glycyl-tRNA synthetase with tRNA supports the synthetase classification and reveals large domain movements.
Sci Adv, 9, 2023
|
|
6L8R
 
 | | membrane-bound PD-L1-CD | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Maorong, W, Cao, Y, Bin, W, Bo, O. | | Deposit date: | 2019-11-07 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PD-L1 degradation is regulated by electrostatic membrane association of its cytoplasmic domain.
Nat Commun, 12, 2021
|
|
8HM4
 
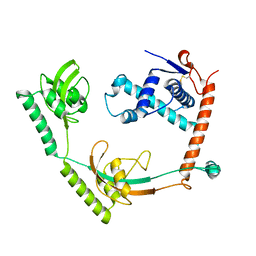 | | Crystal structure of PPIase | | Descriptor: | Peptidylprolyl isomerase | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
8HM3
 
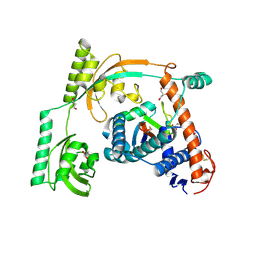 | | Complex of PPIase-BfUbb | | Descriptor: | GLYCEROL, MAGNESIUM ION, Peptidylprolyl isomerase, ... | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
8XHA
 
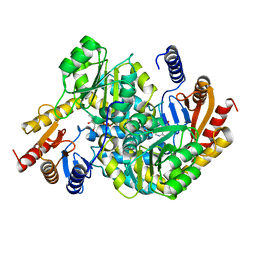 | | Crystal structure of alpha-Oxoamine Synthase Alb29 with PLP cofactor and L-glutamate | | Descriptor: | 8-amino-7-oxononanoate synthase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Xu, M.J, Zhang, D.K. | | Deposit date: | 2023-12-17 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and mechanistic investigations on CC bond forming alpha-oxoamine synthase allowing L-glutamate as substrate.
Int.J.Biol.Macromol., 268, 2024
|
|
8XHD
 
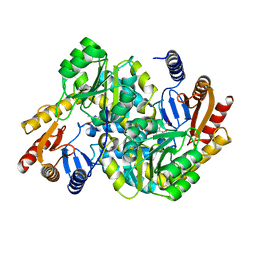 | | Crystal structure of alpha-Oxoamine Synthase Alb29 with PLP cofactor and L-glutamate | | Descriptor: | 8-amino-7-oxononanoate synthase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Xu, M.J, Zhang, D.K. | | Deposit date: | 2023-12-17 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and mechanistic investigations on CC bond forming alpha-oxoamine synthase allowing L-glutamate as substrate.
Int.J.Biol.Macromol., 268, 2024
|
|
8XHK
 
 | | Crystal structure of alpha-Oxoamine Synthase Alb29 with PLP cofactor | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Xu, M.J, Zhang, D.K. | | Deposit date: | 2023-12-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and mechanistic investigations on CC bond forming alpha-oxoamine synthase allowing L-glutamate as substrate.
Int.J.Biol.Macromol., 268, 2024
|
|
6X3P
 
 | | Co-structure of BTK kinase domain with L-005298385 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-{8-amino-3-[(6R,8aS)-3-oxooctahydroindolizin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-3-(cyclopropyloxy)-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6X3N
 
 | | Co-structure of BTK kinase domain with L-005085737 inhibitor | | Descriptor: | 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-{8-amino-3-[(6R,8aS)-3-oxohexahydro-3H-[1,3]oxazolo[3,4-a]pyridin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6X3O
 
 | | Co-structure of BTK kinase domain with L-005191930 inhibitor | | Descriptor: | 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-{8-amino-3-[(6R,8aS)-3-oxo-3,5,6,7,8,8a-hexahydroindolizin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-3-methoxy-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
