4UTQ
 
 | | A structural model of the active ribosome-bound membrane protein insertase YidC | | Descriptor: | ATP SYNTHASE SUBUNIT C, MEMBRANE PROTEIN INSERTASE YIDC | | Authors: | Wickles, S, Singharoy, A, Andreani, J, Seemayer, S, Bischoff, L, Berninghausen, O, Soeding, J, Schulten, K, vanderSluis, E.O, Beckmann, R. | | Deposit date: | 2014-07-22 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A Structural Model of the Active Ribosome-Bound Membrane Protein Insertase Yidc.
Elife, 3, 2014
|
|
1BLX
 
 | | P19INK4D/CDK6 COMPLEX | | Descriptor: | CALCIUM ION, CYCLIN-DEPENDENT KINASE 6, P19INK4D | | Authors: | Brotherton, D.H, Dhanaraj, V, Wick, S, Brizuela, L, Domaille, P.J, Volyanik, E, Xu, X, Parisini, E, Smith, B.O, Archer, S.J, Serrano, M, Brenner, S.L, Blundell, T.L, Laue, E.D. | | Deposit date: | 1998-07-21 | | Release date: | 1999-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d.
Nature, 395, 1998
|
|
4DKE
 
 | | Crystal Structure of Human Interleukin-34 Bound to FAb1.1 | | Descriptor: | FAb1.1 Heavy Chain, FAb1.1 Light Chain, Interleukin-34, ... | | Authors: | Ma, X, Chen, Y, Stawicki, S, Wu, Y, Bazan, J.F, Starovasnik, M.A. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for the Dual Recognition of Helical Cytokines IL-34 and CSF-1 by CSF-1R.
Structure, 20, 2012
|
|
4DKF
 
 | | Crystal Structure of Human Interleukin-34 Bound to FAb2 | | Descriptor: | FAb2 Heavy Chain, FAb2 Light Chain, Interleukin-34, ... | | Authors: | Ma, X, Chen, Y, Stawicki, S, Wu, Y, Bazan, J.F, Starovasnik, M.A. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for the Dual Recognition of Helical Cytokines IL-34 and CSF-1 by CSF-1R.
Structure, 20, 2012
|
|
8GKV
 
 | | Crystal structure of anti-adaptor IraP that regulates RpoS proteolysis | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Shaw, G.X, Gan, J, Suburaman, P, Battesti, A, Zhou, Y.N, Wickner, S, Gottesman, S, Ji, X. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structural and functional study of anti-adaptor IraP-mediated regulation of RpoS proteolysis
to be published
|
|
3J16
 
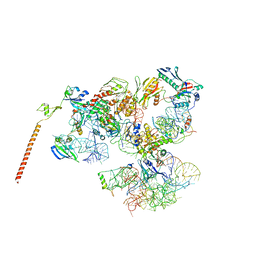 | | Models of ribosome-bound Dom34p and Rli1p and their ribosomal binding partners | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S24-A, ... | | Authors: | Becker, T, Franckenberg, S, Wickles, S, Shoemaker, C.J, Anger, A.M, Armache, J.-P, Sieber, H, Ungewickell, C, Berninghausen, O, Daberkow, I, Karcher, A, Thomm, M, Hopfner, K.-P, Green, R, Beckmann, R. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural basis of highly conserved ribosome recycling in eukaryotes and archaea.
Nature, 482, 2012
|
|
3J15
 
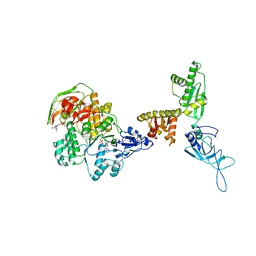 | | Model of ribosome-bound archaeal Pelota and ABCE1 | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, ... | | Authors: | Becker, T, Franckenberg, S, Wickles, S, Shoemaker, C.J, Anger, A.M, Armache, J.-P, Sieber, H, Ungewickell, C, Berninghausen, O, Daberkow, I, Karcher, A, Thomm, M, Hopfner, K.-P, Green, R, Beckmann, R. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis of highly conserved ribosome recycling in eukaryotes and archaea.
Nature, 482, 2012
|
|
3J25
 
 | | Structural basis for TetM-mediated tetracycline resistance | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Tetracycline resistance protein tetM | | Authors: | Doenhoefer, A, Franckenberg, S, Wickles, S, Berninghausen, O, Beckmann, R, Wilson, D.N. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural basis for TetM-mediated tetracycline resistance.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5ABB
 
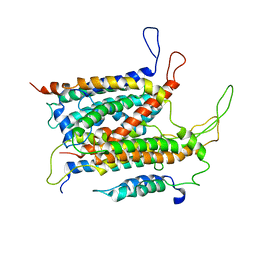 | | Visualization of a polytopic membrane protein during SecY-mediated membrane insertion | | Descriptor: | GREEN-LIGHT ABSORBING PROTEORHODOPSIN, PROTEIN TRANSLOCASE SUBUNIT SECE, PROTEIN TRANSLOCASE SUBUNIT SECY | | Authors: | Bischoff, L, Wickles, S, Berninghausen, O, vanderSluis, E, Beckmann, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Visualization of a Polytopic Membrane Protein During Secy-Mediated Membrane Insertion.
Nat.Commun., 5, 2014
|
|
5A7U
 
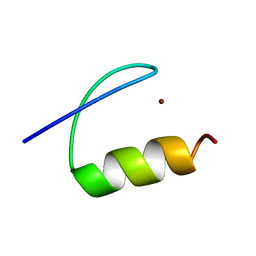 | | Single-particle cryo-EM of co-translational folded adr1 domain inside the E. coli ribosome exit tunnel. | | Descriptor: | REGULATORY PROTEIN ADR1, ZINC ION | | Authors: | Nilsson, O.B, Hedman, R, Marino, J, Wickles, S, Bischoff, L, Johansson, M, Muller-Lucks, A, Trovato, F, Puglisi, J.D, O'Brien, E, Beckmann, R, von Heijne, G. | | Deposit date: | 2015-07-10 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cotranslational Protein Folding Inside the Ribosome Exit Tunnel.
Cell Rep., 12, 2015
|
|
5M5H
 
 | | RIBOSOME-BOUND YIDC INSERTASE | | Descriptor: | Membrane protein insertase YidC | | Authors: | Kedrov, A, Wickles, S, Crevenna, A.H, van der Sluis, E, Buschauer, R, Berninghausen, O, Lamb, D.C, Beckmann, R. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Dynamics of the YidC:Ribosome Complex during Membrane Protein Biogenesis.
Cell Rep, 17, 2016
|
|
2MHZ
 
 | | Structure of Exocyclic S,S N6,N6-(2,3-Dihydroxy-1,4-butadiyl)-2'-Deoxyadenosine Adduct Induced by 1,2,3,4-Diepoxybutane in DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(SDE)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Kowal, E.A, Seneviratne, U, Wickramaratne, S, Doherty, K.E, Cao, X, Tretyakova, N, Stone, M.P. | | Deposit date: | 2013-12-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Exocyclic R,R- and S,S-N(6),N(6)-(2,3-Dihydroxybutan-1,4-diyl)-2'-Deoxyadenosine Adducts Induced by 1,2,3,4-Diepoxybutane.
Chem.Res.Toxicol., 27, 2014
|
|
2MHX
 
 | | Structure of Exocyclic R,R N6,N6-(2,3-Dihydroxy-1,4-butadiyl)-2'-Deoxyadenosine Adduct Induced by 1,2,3,4-Diepoxybutane in DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(RBD)P*AP*GP*AP*AP*G)-3'), 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3') | | Authors: | Kowal, E.A, Seneviratne, U, Wickramaratne, S, Doherty, K.E, Cao, X, Tretyakova, N, Stone, M.P. | | Deposit date: | 2013-12-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Exocyclic R,R- and S,S-N(6),N(6)-(2,3-Dihydroxybutan-1,4-diyl)-2'-Deoxyadenosine Adducts Induced by 1,2,3,4-Diepoxybutane.
Chem.Res.Toxicol., 27, 2014
|
|
4GDX
 
 | | Crystal Structure of Human Gamma-Glutamyl Transpeptidase--Glutamate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | West, M.B, Chen, Y, Wickham, S, Heroux, A, Cahill, K, Hanigan, M.H, Mooers, B.H.M. | | Deposit date: | 2012-08-01 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Novel Insights into Eukaryotic gamma-Glutamyltranspeptidase 1 from the Crystal Structure of the Glutamate-bound Human Enzyme.
J.Biol.Chem., 288, 2013
|
|
4GG2
 
 | | The crystal structure of glutamate-bound human gamma-glutamyltranspeptidase 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | West, M.B, Chen, Y, Wickham, S, Heroux, A, Cahill, K, Hanigan, M.H, Mooers, B.H.M. | | Deposit date: | 2012-08-04 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Novel Insights into Eukaryotic gamma-Glutamyltranspeptidase 1 from the Crystal Structure of the Glutamate-bound Human Enzyme.
J.Biol.Chem., 288, 2013
|
|
6J13
 
 | | Redox protein from Chlamydomonas reinhardtii | | Descriptor: | 2-cys peroxiredoxin | | Authors: | Charoenwattansatien, R, Zinzius, K, Tanaka, H, Hippler, M, Kurisu, G. | | Deposit date: | 2018-12-27 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Calcium sensing via EF-hand 4 enables thioredoxin activity in the sensor-responder protein calredoxin in the green algaChlamydomonas reinhardtii.
J.Biol.Chem., 295, 2020
|
|
8CBM
 
 | | Structure of human mitochondrial CCA-adding enzyme in complex with mitochondrial pre-tRNA-Ile | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase type-2, CCA tRNA nucleotidyltransferase 1, mitochondrial, ... | | Authors: | MEYNIER, V, HARDWICK, S, CATALA, M, ROSKE, J, OERUM, S, CHIRGADZE, D, BARRAUD, P, LUISI, B, TISNE, C. | | Deposit date: | 2023-01-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for human mitochondrial tRNA maturation.
Nat Commun, 15, 2024
|
|
8CBO
 
 | | Structure of human mitochondrial MRPP1-MRPP2 in complex with mitochondrial pre-tRNA-Ile | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase type-2, Mitochondrial Precursor tRNA-Ile(5,4), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | MEYNIER, V, HARDWICK, S, CATALA, M, ROSKE, J, OERUM, S, CHIRGADZE, D, BARRAUD, P, LUISI, B, TISNE, C. | | Deposit date: | 2023-01-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for human mitochondrial tRNA maturation.
Nat Commun, 15, 2024
|
|
8CBK
 
 | | Structure of human mitochondrial RNase P in complex with mitochondrial pre-tRNA-His(5,Ser) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase type-2, MAGNESIUM ION, Mitochondrial Precursor tRNA-His(5,Ser), ... | | Authors: | MEYNIER, V, HARDWICK, S, CATALA, M, ROSKE, J, OERUM, S, CHIRGADZE, D, BARRAUD, P, LUISI, B, TISNE, C. | | Deposit date: | 2023-01-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis for human mitochondrial tRNA maturation.
Nat Commun, 15, 2024
|
|
8CBL
 
 | | Structure of human mitochondrial RNase Z in complex with mitochondrial pre-tRNA-His(0,Ser) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase type-2, Mitochondrial Precursor tRNA-His(0,Ser), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | MEYNIER, V, HARDWICK, S, CATALA, M, ROSKE, J, OERUM, S, CHIRGADZE, D, BARRAUD, P, YU, W, LUISI, B, TISNE, C. | | Deposit date: | 2023-01-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural basis for human mitochondrial tRNA maturation.
Nat Commun, 15, 2024
|
|
6F62
 
 | |
7NZM
 
 | | Cryo-EM structure of pre-dephosphorylation complex of phosphorylated eIF2alpha with trapped holophosphatase (PP1A_D64A/PPP1R15A/G-actin/DNase I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yan, Y, Hardwick, S, Ron, D. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Higher-order phosphatase-substrate contacts terminate the integrated stress response.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5NB9
 
 | | Structure of the N-terminal domain of the Escherichia Coli ProQ RNA binding protein | | Descriptor: | RNA chaperone ProQ | | Authors: | Gonzales, G, Hardwick, S, Maslen, S, Skehel, M, Holmqvist, E, Vogel, J, Bateman, A, Luisi, B, Broadhurst, R. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of the Escherichia coli ProQ RNA-binding protein.
RNA, 23, 2017
|
|
5NBB
 
 | | Structure of the C-terminal domain of the Escherichia Coli ProQ RNA binding protein | | Descriptor: | RNA chaperone ProQ | | Authors: | Gonzales, G, Hardwick, S, Maslen, S, Skehel, M, Holmqvist, E, Vogel, J, Bateman, A, Luisi, B, Broadhurst, R. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of the Escherichia coli ProQ RNA-binding protein.
RNA, 23, 2017
|
|
6OD1
 
 | | IraD-bound to RssB D58P variant | | Descriptor: | Anti-adapter protein IraD, Regulator of RpoS | | Authors: | Deaconescu, A.M, Dorich, V. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of a response regulator of sigmaSstability by a ClpXP antiadaptor.
Genes Dev., 33, 2019
|
|
