3IQL
 
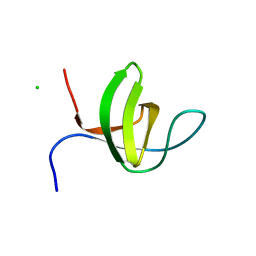 | | Crystal structure of the rat endophilin-A1 SH3 domain | | Descriptor: | CHLORIDE ION, Endophilin-A1 | | Authors: | Trempe, J.F, Kozlov, G, Camacho, E.M, Gehring, K. | | Deposit date: | 2009-08-20 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | SH3 domains from a subset of BAR proteins define a Ubl-binding domain and implicate parkin in synaptic ubiquitination.
Mol.Cell, 36, 2009
|
|
3M3J
 
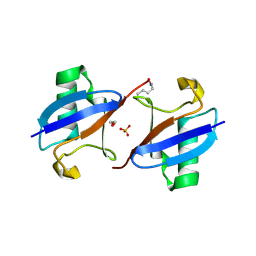 | | A new crystal form of Lys48-linked diubiquitin | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Ubiquitin | | Authors: | Trempe, J.F, Brown, N.R, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new crystal form of Lys48-linked diubiquitin.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
8UDC
 
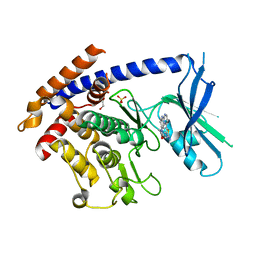 | | Crystal structure of TcPINK1 in complex with CYC116 | | Descriptor: | (4P)-4-(2-amino-4-methyl-1,3-thiazol-5-yl)-N-[4-(morpholin-4-yl)phenyl]pyrimidin-2-amine, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Veyron, S, Rasool, S, Trempe, J.F. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Identification and structural characterization of small molecule inhibitors of PINK1.
Sci Rep, 14, 2024
|
|
8UCT
 
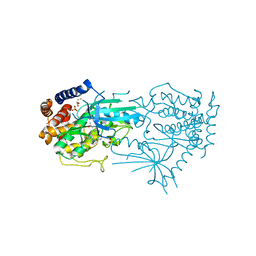 | | Crystal structure of TcPINK1 in complex with PRT | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-{[(1R,2S)-2-aminocyclohexyl]amino}-4-{[3-(2H-1,2,3-triazol-2-yl)phenyl]amino}pyrimidine-5-carboxamide, SULFATE ION, ... | | Authors: | Veyron, S, Rasool, S, Trempe, J.F. | | Deposit date: | 2023-09-27 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Characterization of a new family of PINK1 inhibitors
To Be Published
|
|
6UCC
 
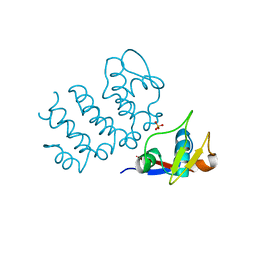 | | Structure of human PACRG-MEIG1 complex (limited proteolysis) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Meiosis expressed gene 1 protein homolog, PHOSPHATE ION, ... | | Authors: | Khan, N, Croteau, N, Pelletier, D, Veyron, S, Trempe, J.F. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human PACRG in complex with MEIG1
Biorxiv, 2019
|
|
4ZYN
 
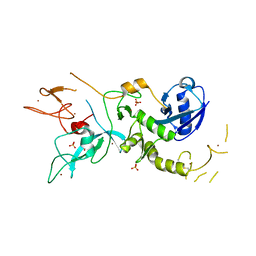 | | Crystal Structure of Parkin E3 ubiquitin ligase (linker deletion; delta 86-130) | | Descriptor: | E3 ubiquitin-protein ligase parkin, SULFATE ION, ZINC ION | | Authors: | Lilov, A, Sauve, V, Trempe, J.F, Rodionov, D, Wang, J, Gehring, K. | | Deposit date: | 2015-05-21 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | A Ubl/ubiquitin switch in the activation of Parkin.
Embo J., 34, 2015
|
|
7MP8
 
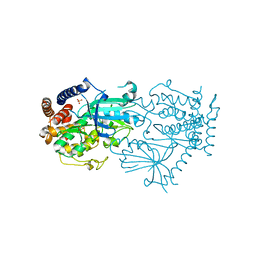 | |
7MP9
 
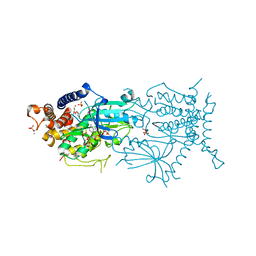 | | Crystal structure of the cytosolic domain of Tribolium castaneum PINK1 phosphorylated at Ser205 in complex with ADP analog | | Descriptor: | AMP PHOSPHORAMIDATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Rasool, S, Veyron, S, Trempe, J.F. | | Deposit date: | 2021-05-04 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of PINK1 activation by autophosphorylation and insights into assembly on the TOM complex.
Mol.Cell, 82, 2022
|
|
6DJW
 
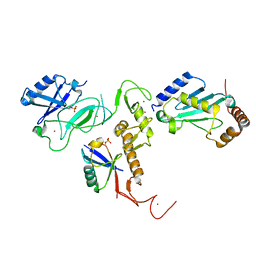 | | Crystal Structure of pParkin (REP and RING2 deleted)-pUb-UbcH7 complex | | Descriptor: | RBR-type E3 ubiquitin transferase,RBR-type E3 ubiquitin transferase, Ubiquitin, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Sauve, V, Sung, G, Trempe, J.F, Gehring, K. | | Deposit date: | 2018-05-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Mechanism of parkin activation by phosphorylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DJX
 
 | | Crystal Structure of pParkin-pUb-UbcH7 complex | | Descriptor: | RBR-type E3 ubiquitin transferase,RBR-type E3 ubiquitin transferase, Ubiquitin, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Sauve, V, Sung, G, Trempe, J.F, Gehring, K. | | Deposit date: | 2018-05-27 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.801 Å) | | Cite: | Mechanism of parkin activation by phosphorylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6NEP
 
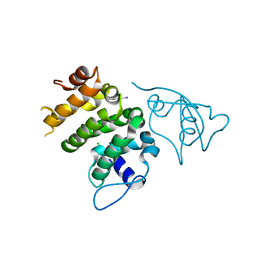 | | Structure of human PACRG-MEIG1 complex | | Descriptor: | CHLORIDE ION, Meiosis expressed gene 1 protein homolog, PHOSPHATE ION, ... | | Authors: | Khan, N, Croteau, N, Pelletier, D, Veyron, S, Trempe, J.F. | | Deposit date: | 2018-12-18 | | Release date: | 2019-10-23 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.097625 Å) | | Cite: | Crystal structure of human PACRG in complex with MEIG1
Biorxiv, 2019
|
|
6NDU
 
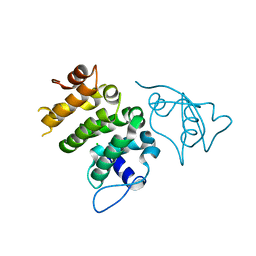 | | Structure of human PACRG-MEIG1 complex | | Descriptor: | CHLORIDE ION, Meiosis expressed gene 1 protein homolog, PHOSPHATE ION, ... | | Authors: | Khan, N, Croteau, N, Pelletier, D, Veyron, S, Trempe, J.F. | | Deposit date: | 2018-12-14 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human PACRG in complex with MEIG1
Biorxiv, 2019
|
|
1NEE
 
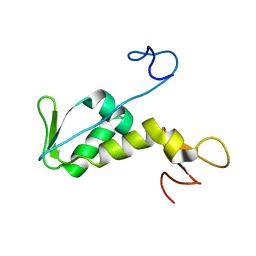 | | Structure of archaeal translation factor aIF2beta from Methanobacterium thermoautrophicum | | Descriptor: | Probable translation initiation factor 2 beta subunit, ZINC ION | | Authors: | Gutierrez, P, Trempe, J.F, Siddiqui, N, Arrowsmith, C, Gehring, K. | | Deposit date: | 2002-12-11 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the archaeal translation initiation factor aIF2beta from Methanobacterium thermoautotrophicum: Implications for translation initiation.
Protein Sci., 13, 2004
|
|
1NMR
 
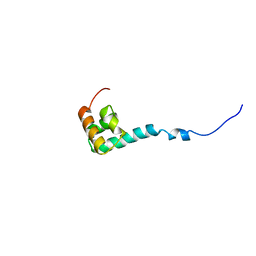 | | Solution Structure of C-terminal Domain from Trypanosoma cruzi Poly(A)-Binding Protein | | Descriptor: | poly(A)-binding protein | | Authors: | Siddiqui, N, Kozlov, G, D'Orso, I, Trempe, J.F, Frasch, A.C.C, Gehring, K. | | Deposit date: | 2003-01-10 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Domain from poly(A)-binding protein in Trypanosoma cruzi: A vegetal PABC domain
Protein Sci., 12, 2003
|
|
1FC8
 
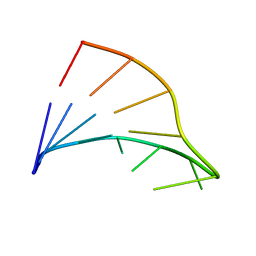 | |
5KES
 
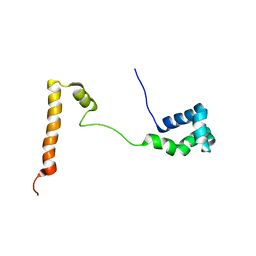 | | Solution structure of the yeast Ddi1 HDD domain | | Descriptor: | DNA damage-inducible protein 1 | | Authors: | Trempe, J.-F, Ratcliffe, C, Veverka, V, Saskova, K, Gehring, K. | | Deposit date: | 2016-06-10 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies of the yeast DNA damage-inducible protein Ddi1 reveal domain architecture of this eukaryotic protein family.
Sci Rep, 6, 2016
|
|
4Z2Z
 
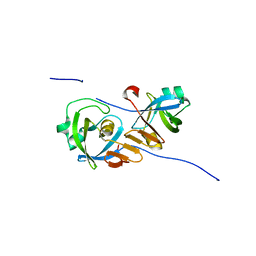 | |
4K95
 
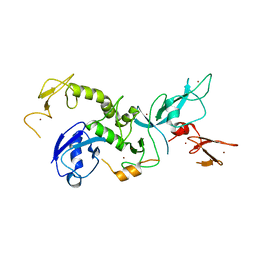 | | Crystal Structure of Parkin | | Descriptor: | E3 ubiquitin-protein ligase parkin, ZINC ION | | Authors: | Seirafi, M, Menade, M, Sauve, V, Kozlov, G, Trempe, J.-F, Nagar, B, Gehring, K. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.499 Å) | | Cite: | Structure of parkin reveals mechanisms for ubiquitin ligase activation.
Science, 340, 2013
|
|
4K7D
 
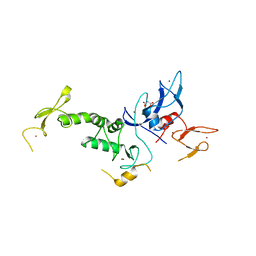 | | Crystal Structure of Parkin C-terminal RING domains | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, MALONATE ION, ... | | Authors: | Sauve, V, Trempe, J.-F, Menade, M, Gehring, K. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of parkin reveals mechanisms for ubiquitin ligase activation.
Science, 340, 2013
|
|
2KNB
 
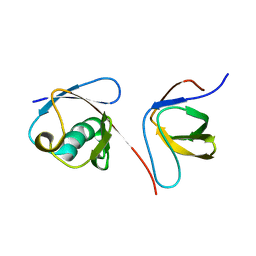 | | Solution NMR structure of the parkin Ubl domain in complex with the endophilin-A1 SH3 domain | | Descriptor: | E3 ubiquitin-protein ligase parkin, Endophilin-A1 | | Authors: | Trempe, J, Guennadi, K, Edna, C.M, Kalle, G. | | Deposit date: | 2009-08-20 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | SH3 domains from a subset of BAR proteins define a Ubl-binding domain and implicate parkin in synaptic ubiquitination.
Mol.Cell, 36, 2009
|
|
2N7E
 
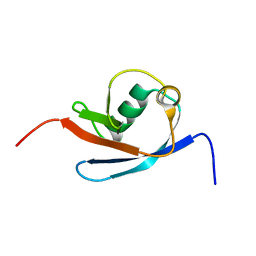 | |
2X5N
 
 | | Crystal Structure of the SpRpn10 VWA domain | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN10, SULFATE ION | | Authors: | Riedinger, C, Boehringer, J, Trempe, J.-F, Lowe, E.D, Brown, N.R, Gehring, K, Noble, M.E.M, Gordon, C, Endicott, J.A. | | Deposit date: | 2010-02-10 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of Rpn10 and its Interactions with Polyubiquitin Chains and the Proteasome Subunit Rpn12.
J.Biol.Chem., 285, 2010
|
|
6NBQ
 
 | | T.elongatus NDH (data-set 1) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 2, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-09 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
6NBX
 
 | | T.elongatus NDH (data-set 2) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 1, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
6NBY
 
 | | T.elongatus NDH (composite model) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 1, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-02-27 | | Last modified: | 2020-04-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
