8CTQ
 
 | | Crystal structure of engineered phospholipase D mutant superPLD 2-48 | | Descriptor: | PHOSPHATE ION, Phospholipase D | | Authors: | Tei, R, Bagde, S.R, Fromme, J.C, Baskin, J.M. | | Deposit date: | 2022-05-16 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Activity-based directed evolution of a membrane editor in mammalian cells.
Nat.Chem., 15, 2023
|
|
8CTP
 
 | | Crystal structure of engineered phospholipase D mutant superPLD 2-23 | | Descriptor: | PHOSPHATE ION, Phospholipase D | | Authors: | Tei, R, Bagde, S.R, Fromme, J.C, Baskin, J.M. | | Deposit date: | 2022-05-16 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activity-based directed evolution of a membrane editor in mammalian cells.
Nat.Chem., 15, 2023
|
|
5C7X
 
 | | Crystal structure of MOR04357, a neutralizing anti-human GM-CSF antibody Fab fragment in complex with human GM-CSF | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, DI(HYDROXYETHYL)ETHER, Granulocyte-macrophage colony-stimulating factor, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
5D71
 
 | | Crystal structure of MOR04302, a neutralizing anti-human GM-CSF antibody Fab fragment in complex with human GM-CSF | | Descriptor: | DI(HYDROXYETHYL)ETHER, Granulocyte-macrophage colony-stimulating factor, Immunglobulin G1 Fab fragment, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-08-13 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
5D70
 
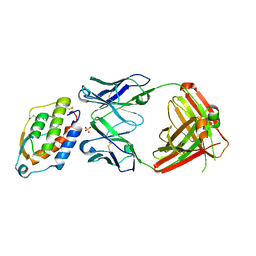 | | Crystal structure of MOR03929, a neutralizing anti-human GM-CSF antibody Fab fragment in complex with human GM-CSF | | Descriptor: | Granulocyte-macrophage colony-stimulating factor, Immunglobulin G1 Fab fragment, heavy chain, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-08-13 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
5D7S
 
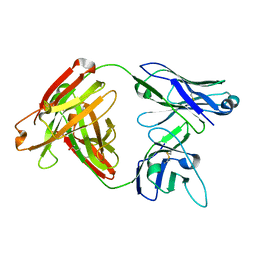 | | Crystal structure of MOR04357, a neutralizing anti-human GM-CSF antibody Fab fragment | | Descriptor: | (2S,3S)-butane-2,3-diol, Immunglobulin G1 Fab fragment, heavy chain, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-08-14 | | Release date: | 2015-10-14 | | Last modified: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
5D72
 
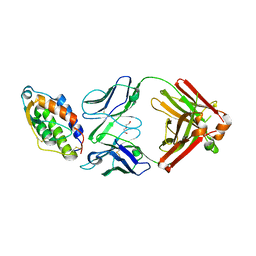 | | Crystal structure of MOR04252, a neutralizing anti-human GM-CSF antibody Fab fragment in complex with human GM-CSF | | Descriptor: | DI(HYDROXYETHYL)ETHER, Granulocyte-macrophage colony-stimulating factor, Immunglobulin G1 Fab fragment, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-08-13 | | Release date: | 2015-10-14 | | Last modified: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
7Z0T
 
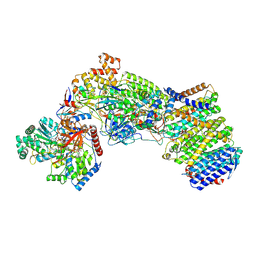 | | Structure of the Escherichia coli formate hydrogenlyase complex (aerobic preparation, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CARBONMONOXIDE-(DICYANO) IRON, FE (III) ION, ... | | Authors: | Steinhilper, R, Murphy, B.J. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-28 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the membrane-bound formate hydrogenlyase complex from Escherichia coli.
Nat Commun, 13, 2022
|
|
7Z0S
 
 | |
3RBG
 
 | | Crystal structure analysis of Class-I MHC restricted T-cell associated molecule | | Descriptor: | Cytotoxic and regulatory T-cell molecule, PHOSPHATE ION | | Authors: | Rubinstein, R, Ramagopal, U.A, Toro, R, Nathenson, S.G, Fiser, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2011-03-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional classification of immune regulatory proteins.
Structure, 21, 2013
|
|
1PYI
 
 | |
1D66
 
 | | DNA RECOGNITION BY GAL4: STRUCTURE OF A PROTEIN/DNA COMPLEX | | Descriptor: | CADMIUM ION, DNA (5'-D(*CP*CP*GP*GP*AP*GP*GP*AP*CP*AP*GP*TP*CP*CP*TP*CP*C P*GP*G)-3'), DNA (5'-D(*CP*CP*GP*GP*AP*GP*GP*AP*CP*TP*GP*TP*CP*CP*TP*CP*C P*GP*G)-3'), ... | | Authors: | Marmorstein, R, Carey, M, Ptashne, M, Harrison, S.C. | | Deposit date: | 1992-03-06 | | Release date: | 1992-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | DNA recognition by GAL4: structure of a protein-DNA complex.
Nature, 356, 1992
|
|
1MX4
 
 | | Structure of p18INK4c (F82Q) | | Descriptor: | Cyclin-dependent kinase 6 inhibitor | | Authors: | Marmorstein, R, Venkataramani, R.N, MacLachlan, T.K, Chai, X, El-Deiry, W.S. | | Deposit date: | 2002-10-01 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of p18INK4c proteins with increased thermodynamic stability and cell cycle inhibitory activity
J.Biol.Chem., 277, 2002
|
|
1MX2
 
 | | Structure of F71N mutant of p18INK4c | | Descriptor: | Cyclin-dependent kinase 6 inhibitor | | Authors: | Marmorstein, R, Venkataramani, R.N, MacLachlan, T.K, Chai, X, El-Deiery, W.S. | | Deposit date: | 2002-10-01 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based design of p18INK4c proteins with increased thermodynamic stability and cell cycle inhibitory activity
J.Biol.Chem., 277, 2002
|
|
1MX6
 
 | | Structure of p18INK4c (F92N) | | Descriptor: | Cyclin-dependent kinase 6 inhibitor | | Authors: | Marmorstein, R, Venkataramani, R.N, MacLachlan, T.K, Chai, X, El-Deiry, W.S. | | Deposit date: | 2002-10-01 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of p18INK4c proteins with increased thermodynamic stability and cell cycle inhibitory activity
J.Biol.Chem., 277, 2002
|
|
8PK8
 
 | |
8PKA
 
 | |
8PK9
 
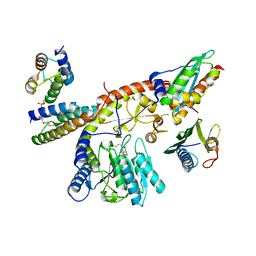 | |
4ZPO
 
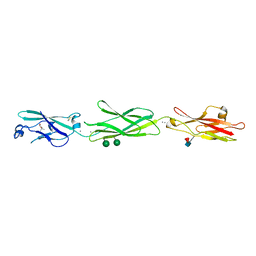 | | Crystal Structure of Protocadherin Gamma C5 EC1-3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MCG133388, ... | | Authors: | Wolcott, H.N, Goodman, K.M, Bahna, F, Mannepalli, S, Rubinstein, R, Honig, B, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
4ZBJ
 
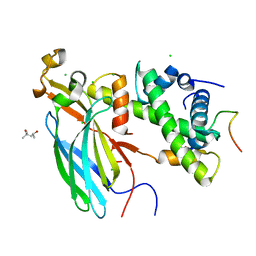 | | UBN1 peptide bound to H3.3/H4/Asf1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Histone H3, ... | | Authors: | Marmorstein, R, Ricketts, M.D, Tang, Y. | | Deposit date: | 2015-04-14 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex.
Nat Commun, 6, 2015
|
|
5T53
 
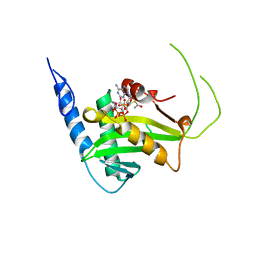 | | MOLECULAR BASIS FOR COHESIN ACETYLATION BY ESTABLISHMENT OF SISTER CHROMATID COHESION N-ACETYLTRANSFERASE ESCO1 | | Descriptor: | ACETYL COENZYME *A, N-acetyltransferase ESCO1, ZINC ION | | Authors: | Marmorstein, R, Rivera-Colon, Y, Liszczak, G.P, Olia, A.S, Maguire, A. | | Deposit date: | 2016-08-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Molecular Basis for Cohesin Acetylation by Establishment of Sister Chromatid Cohesion N-Acetyltransferase ESCO1.
J. Biol. Chem., 291, 2016
|
|
2OD9
 
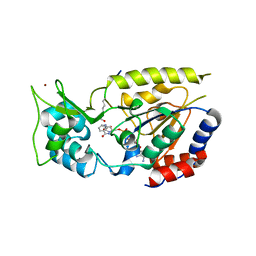 | | Structural Basis for Nicotinamide Inhibition and Base Exchange in Sir2 Enzymes | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, H4 peptide, NAD-dependent deacetylase HST2, ... | | Authors: | Marmorstein, R, Sanders, B.D. | | Deposit date: | 2006-12-21 | | Release date: | 2007-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for nicotinamide inhibition and base exchange in sir2 enzymes.
Mol.Cell, 25, 2007
|
|
2QQF
 
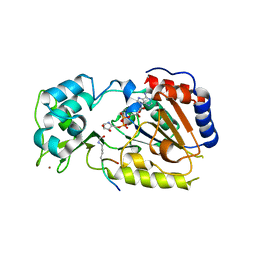 | | Hst2 bound to ADP-HPD and Acetylated histone H4 | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, Histone H4, NAD-dependent deacetylase HST2, ... | | Authors: | Marmorstein, R, Sanders, B.D, Zhao, K, Slama, J. | | Deposit date: | 2007-07-26 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for nicotinamide inhibition and base exchange in sir2 enzymes.
Mol.Cell, 25, 2007
|
|
2QQG
 
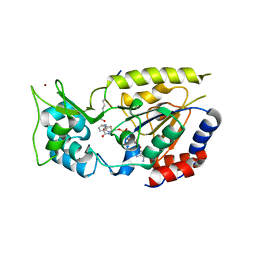 | | Hst2 bound to ADP-HPD, acetyllated histone H4 and nicotinamide | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, Histone H4, NAD-dependent deacetylase HST2, ... | | Authors: | Marmorstein, R, Sanders, B, Zhao, K, Slama, J. | | Deposit date: | 2007-07-26 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for nicotinamide inhibition and base exchange in sir2 enzymes.
Mol.Cell, 25, 2007
|
|
2I32
 
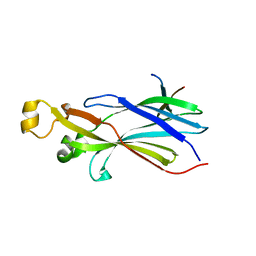 | |
