1QK6
 
 | | Solution structure of huwentoxin-I by NMR | | Descriptor: | HUWENTOXIN-I | | Authors: | Qu, Y, Liang, S, Ding, J, Liu, X, Zhang, R, Gu, X. | | Deposit date: | 1999-07-10 | | Release date: | 1999-08-20 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Proton Nuclear Magnetic Resonance Studies on Huwentoxin-I from the Venom of the Spider Selenocosmia Huwena:2.Three-Dimensional Structure in Solution
J.Protein Chem., 16, 1997
|
|
1RWE
 
 | | Enhancing the activity of insulin at receptor edge: crystal structure and photo-cross-linking of A8 analogues | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z, Xu, B, Chu, Y.C, Li, B, Nakagawa, S.H, Qu, Y, Hu, S.Q, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2003-12-16 | | Release date: | 2005-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enhancing the activity of insulin at the receptor interface: crystal structure and photo-cross-linking of A8 analogues.
Biochemistry, 43, 2004
|
|
5A3C
 
 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes in complex with NAD | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
5A3A
 
 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes (Apo form) | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, SIR2 FAMILY PROTEIN, ... | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
5A3B
 
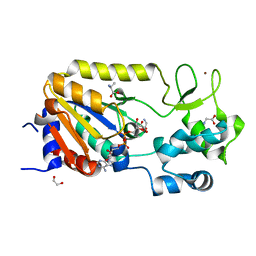 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, ALANINE, ... | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
5A35
 
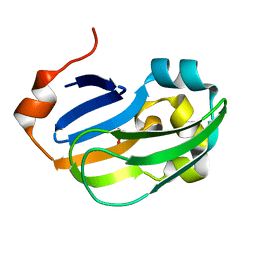 | | Crystal structure of Glycine Cleavage Protein H-Like (GcvH-L) from Streptococcus pyogenes | | Descriptor: | GLYCINE CLEAVAGE SYSTEM H PROTEIN, PENTAETHYLENE GLYCOL | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
8K69
 
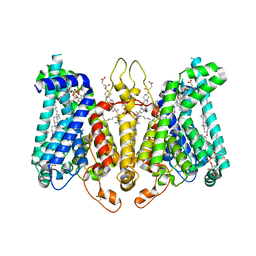 | | Cryo-EM structure of Oryza sativa HKT2;2/1 at 2.3 angstrom | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Wang, X, Shen, X, Qu, Y, Wang, C, Shen, H. | | Deposit date: | 2023-07-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural insights into ion selectivity and transport mechanisms of Oryza sativa HKT2;1 and HKT2;2/1 transporters.
Nat.Plants, 10, 2024
|
|
8K66
 
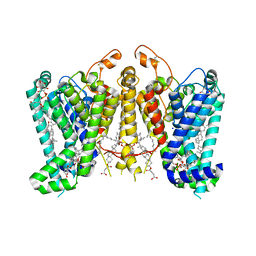 | | Cryo-EM structure of Oryza sativa HKT2;1 at 2.5 angstrom | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Wang, X, Shen, X, Qu, Y, Wang, C, Shen, H. | | Deposit date: | 2023-07-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural insights into ion selectivity and transport mechanisms of Oryza sativa HKT2;1 and HKT2;2/1 transporters.
Nat.Plants, 10, 2024
|
|
7KE5
 
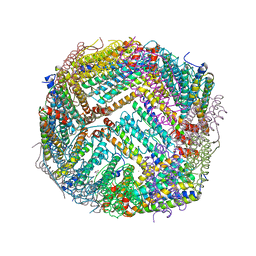 | | Heavy chain ferritin with N-terminal EBNA1 epitope | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Epstein-Barr nuclear antigen 1,Ferritin heavy chain, FE (III) ION | | Authors: | Pederick, J.P, Bruning, J.B. | | Deposit date: | 2020-10-10 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Immunogenicity study of engineered ferritins with C- and N-terminus insertion of Epstein-Barr nuclear antigen 1 epitope.
Vaccine, 39, 2021
|
|
7KE3
 
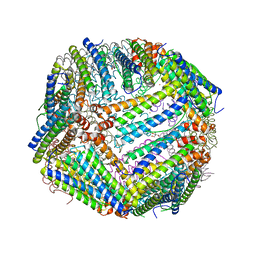 | | Heavy chain ferritin with C-terminal EBNA1 epitope | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, Ferritin heavy chain,Epstein-Barr nuclear antigen 1 | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2020-10-10 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Immunogenicity study of engineered ferritins with C- and N-terminus insertion of Epstein-Barr nuclear antigen 1 epitope.
Vaccine, 39, 2021
|
|
9JQR
 
 | |
9KQK
 
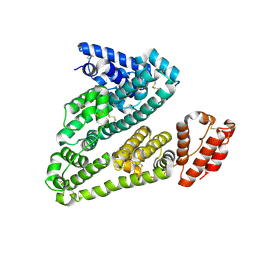 | | Crystal structure of HSA in complex with AmpHecy | | Descriptor: | 2-[[6-[5-[(~{E})-2-[1-[2-[2-(2-methoxyethoxy)ethoxy]ethyl]pyridin-1-ium-4-yl]ethenyl]thiophen-2-yl]naphthalen-2-yl]-methyl-amino]ethanol, Albumin | | Authors: | Chen, X, Shi, M.W, Ge, Y.H, Fang, B, Li, L. | | Deposit date: | 2024-11-26 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Fluorogen-Activating Human Serum Albumin for Mitochondrial Nanoscale Imaging.
Adv Mater, 2025
|
|
9JQP
 
 | |
9JQ6
 
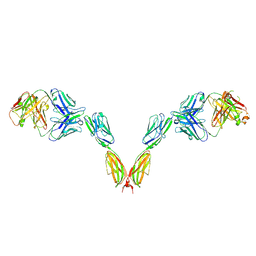 | | Cryo-EM structure of BTN2A1 in complex with antagonist antibody TH002 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BTN2A1 antagonist antibody TH002-Fab heavy chain, ... | | Authors: | Zhang, M, Wang, Y.Q, Xiao, J.Y. | | Deposit date: | 2024-09-27 | | Release date: | 2025-06-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structures of butyrophilin multimers reveal a plier-like mechanism for V gamma 9V delta 2 T cell receptor activation.
Immunity, 2025
|
|
9JQQ
 
 | | Cryo-EM structure of the HMBPP-primed BTN3A1-BTN3A2-BTN2A1 complex | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, M, Wang, Y.Q, Xiao, J.Y. | | Deposit date: | 2024-09-28 | | Release date: | 2025-06-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structures of butyrophilin multimers reveal a plier-like mechanism for V gamma 9V delta 2 T cell receptor activation.
Immunity, 2025
|
|
2F40
 
 | |
1T1K
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT HIS-B10-ASP, VAL-B12-ALA, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin | | Authors: | Huang, K, Xu, B, Hu, S.Q, Chu, Y.C, Hua, Q.X, Whittaker, J, Nakagawa, S.H, De Meyts, P, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-10 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | How Insulin Binds: the B-Chain alpha-Helix Contacts the L1 beta-Helix of the Insulin Receptor.
J.Mol.Biol., 341, 2004
|
|
4J3J
 
 | | Crystal Structure of DPP-IV with Compound C3 | | Descriptor: | Dipeptidyl peptidase 4, N-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butyl]-6-(trifluoromethyl)-3,4-dihydropyrrolo[1,2-a]pyrazine-2(1H)-carboxamide | | Authors: | Xiong, B, Zhu, L.R, Chen, D.Q, Zhao, Y.L, Jiang, F, Shen, J.K. | | Deposit date: | 2013-02-05 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 4-(2,4,5-trifluorophenyl)butane-1,3-diamines as dipeptidyl peptidase IV inhibitors
Chemmedchem, 8, 2013
|
|
1T1Q
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT HIS-B10-ASP, VAL-B12-ABA, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin, insulin | | Authors: | Huang, K, Xu, B, Hu, S.Q, Chu, Y.C, Hua, Q.X, Whittaker, J, Nakagawa, S.H, De Meyts, P, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-10 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | How Insulin Binds: the B-Chain alpha-Helix Contacts the L1 beta-Helix of the Insulin Receptor.
J.Mol.Biol., 341, 2004
|
|
1T1P
 
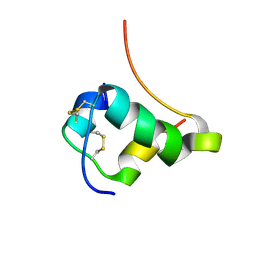 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT HIS-B10-ASP, VAL-B12-THR, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin, insulin | | Authors: | Huang, K, Xu, B, Hu, S.Q, Chu, Y.C, Hua, Q.X, Whittaker, J, Nakagawa, S.H, De Meyts, P, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-10 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | How Insulin Binds: the B-Chain alpha-Helix Contacts the L1 beta-Helix of the Insulin Receptor.
J.Mol.Biol., 341, 2004
|
|
9IS8
 
 | | Cryo-EM structure of AKT1-AtKC1(G315D) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium channel AKT1, ... | | Authors: | Dongliang, L, Zijie, Z, Yannan, Q, Yuyue, T, Huaizong, S. | | Deposit date: | 2024-07-17 | | Release date: | 2025-05-21 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | AtKC1 inhibits AKT1 activation via its amino-terminal inhibitory domain.
Plant Commun., 6, 2025
|
|
7XUX
 
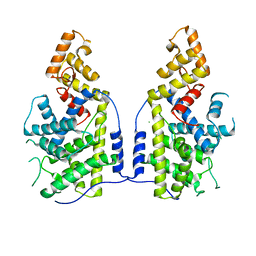 | |
7XTV
 
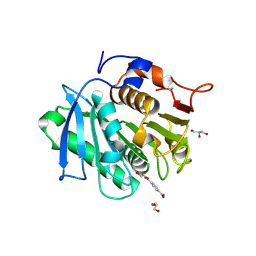 | | The structure of MHET-bound TfCut S130A | | Descriptor: | 4-(2-hydroxyethyloxycarbonyl)benzoic acid, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTU
 
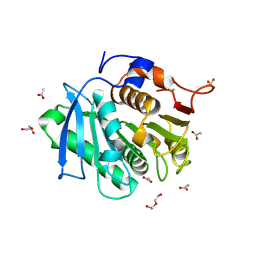 | | The structure of TfCut S130A | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTS
 
 | | The apo structure of the engineered TfCut S130A | | Descriptor: | SODIUM ION, alpha/beta hydrolase | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
