2PFD
 
 | | Anisotropically refined structure of FTCD | | Descriptor: | Formimidoyltransferase-cyclodeaminase | | Authors: | Poon, B.K, Chen, X, Lu, M, Quiocho, F.A, Wang, Q, Ma, J. | | Deposit date: | 2007-04-04 | | Release date: | 2007-04-24 | | Last modified: | 2011-08-10 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Anisotropically refined structure of FTCD
To be Published
|
|
2QTO
 
 | | An anisotropic model for potassium channel KcsA | | Descriptor: | POTASSIUM ION, Voltage-gated potassium channel | | Authors: | Chen, X, Poon, B.K, Dousis, A, Wang, Q, Ma, J. | | Deposit date: | 2007-08-02 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Normal-mode refinement of anisotropic thermal parameters for potassium channel KcsA at 3.2 A crystallographic resolution
Structure, 15, 2007
|
|
7OUI
 
 | | Structure of C2S2M2-type Photosystem supercomplex from Arabidopsis thaliana (digitonin-extracted) | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Graca, A.T, Hall, M, Persson, K, Schroder, W.P. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | High-resolution model of Arabidopsis Photosystem II reveals the structural consequences of digitonin-extraction.
Sci Rep, 11, 2021
|
|
3FUS
 
 | | Improved Structure of the Unliganded Simian Immunodeficiency Virus gp120 Core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, X, Poon, B, Wang, Q, Ma, J. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural improvement of unliganded simian immunodeficiency virus gp120 core by normal-mode-based X-ray crystallographic refinement.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6Y6K
 
 | | Cryo-EM structure of a Phenuiviridae L protein | | Descriptor: | MAGNESIUM ION, RNA-dependent RNA polymerase | | Authors: | Vogel, D, Thorkelsson, S.R, Quemin, E, Meier, K, Kouba, T, Gogrefe, N, Busch, C, Reindl, S, Guenther, S, Cusack, S, Gruenewald, K, Rosenthal, M. | | Deposit date: | 2020-02-26 | | Release date: | 2020-04-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural and functional characterization of the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 48, 2020
|
|
8G9B
 
 | | Human IMPDH2 mutant - L245P, treated with GTP, ATP, IMP, and NAD+; compressed filament segment reconstruction | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | O'Neill, A.G, Kollman, J.M. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
8G8F
 
 | | Human IMPDH2 mutant - L245P, treated with ATP, IMP, and NAD+; extended filament segment reconstruction | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | O'Neill, A.G, Kollman, J.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
7LM4
 
 | | The crystal structure of the I38T mutant PA Endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988503 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7BJ3
 
 | | ScpA from Streptococcus pyogenes, S512A active site mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | Authors: | Kagawa, T.F, O'Connell, M.R, Cooney, J.C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Enzyme kinetic and binding studies identify determinants of specificity for the immunomodulatory enzyme ScpA, a C5a inactivating bacterial protease.
Comput Struct Biotechnol J, 19, 2021
|
|
7BBM
 
 | | Mutant nitrobindin M75L/H76L/Q96C/M148L (NB4H) from Arabidopsis thaliana with cofactor MnPPIX | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE PROTOPORPHYRIN IX, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Minges, A, Sauer, D.F, Wittwer, M, Markel, U, Spiertz, M, Schiffels, J, Davari, M.D, Okuda, J, Schwaneberg, U, Groth, G. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Chemogenetic engineering of nitrobindin toward an artificial epoxygenase
Catalysis Science And Technology, 2021
|
|
8TJG
 
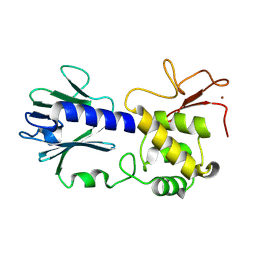 | |
8TUA
 
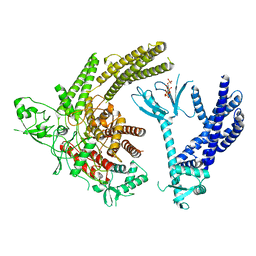 | | Full-length P-Rex1 in complex with inositol 1,3,4,5-tetrakisphosphate (IP4) | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2023-08-15 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Full-length P-Rex1 in complex with inositol 1,3,4,5-tetrakisphosphate (IP4)
Elife, 2024
|
|
7ZJ5
 
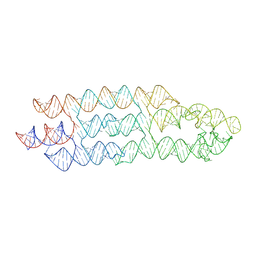 | | Unbound state of a brocolli-pepper aptamer FRET tile. | | Descriptor: | POTASSIUM ION, brocolli-pepper aptamer | | Authors: | McRae, E.K.S, Vallina, N.S, Hansen, B.K, Boussebayle, A, Andersen, E.S. | | Deposit date: | 2022-04-08 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structure determination of Pepper-Broccoli FRET pair by RNA origami scaffolding
To Be Published
|
|
7ZJ4
 
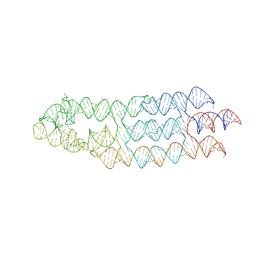 | | Ligand bound state of a brocolli-pepper aptamer FRET tile | | Descriptor: | 4-(3,5-difluoro-4-hydroxybenzyl)-1,2-dimethyl-1H-imidazol-5-ol, 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]thieno[3,2-b]thiophen-2-yl]ethenyl]benzenecarbonitrile, POTASSIUM ION, ... | | Authors: | McRae, E.K.S, Vallina, N.S, Hansen, B.K, Boussebayle, A, Andersen, E.S. | | Deposit date: | 2022-04-08 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structure determination of Pepper-Broccoli FRET pair by RNA origami scaffolding
To Be Published
|
|
7OS2
 
 | | Cryo-EM structure of Brr2 in complex with Jab1/MPN and C9ORF78 | | Descriptor: | Pre-mRNA-processing-splicing factor 8, Telomere length and silencing protein 1 homolog, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Bergfort, A, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-06-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
7OS1
 
 | | Cryo-EM structure of Brr2 in complex with Fbp21 | | Descriptor: | U5 small nuclear ribonucleoprotein 200 kDa helicase, WW domain-binding protein 4 | | Authors: | Bergfort, A, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-06-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
7PV9
 
 | |
7PPN
 
 | | SHP2 catalytic domain in complex with CD28 (183-198) phosphopeptide (pTyr-191, p-Thr-195) | | Descriptor: | GLYCEROL, T-cell-specific surface glycoprotein CD28, Tyrosine-protein phosphatase non-receptor type 11,Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sok, P, Zeke, A, Remenyi, A. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling.
Nat Commun, 13, 2022
|
|
7PPM
 
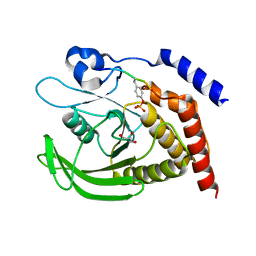 | | SHP2 catalytic domain in complex with IRS1 (889-901) phosphopeptide (pSer-892, pTyr-896) | | Descriptor: | GLYCEROL, Insulin receptor substrate 1, Tyrosine-protein phosphatase non-receptor type 11,Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sok, P, Zeke, A, Remenyi, A. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling.
Nat Commun, 13, 2022
|
|
7PPL
 
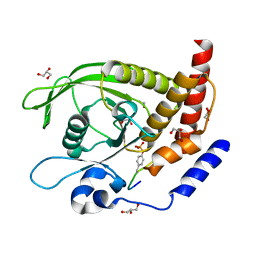 | | SHP2 catalytic domain in complex with IRS1 (625-639) phosphopeptide (pTyr-632, pSer-636) | | Descriptor: | ETHANOL, GLYCEROL, Insulin receptor substrate 1, ... | | Authors: | Sok, P, Zeke, A, Remenyi, A. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling.
Nat Commun, 13, 2022
|
|
7PTH
 
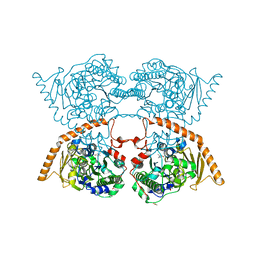 | |
7PJH
 
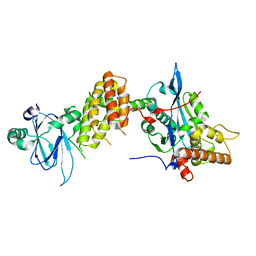 | | Crystal structure of the human spliceosomal maturation factor AAR2 bound to the RNAse H domain of PRPF8 | | Descriptor: | Pre-mRNA-processing-splicing factor 8, Protein AAR2 homolog | | Authors: | Preussner, M, Santos, K, Heroven, A.C, Alles, J, Heyd, F, Wahl, M.C, Weber, G. | | Deposit date: | 2021-08-24 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional investigation of the human snRNP assembly factor AAR2 in complex with the RNase H-like domain of PRPF8.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PUX
 
 | | Structure of p97 N-D1(L198W) in complex with Fragment TROLL2 | | Descriptor: | (1S)-2-amino-1-(4-bromophenyl)ethan-1-ol, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bothe, S, Schindelin, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Fragment screening using biolayer interferometry reveals ligands targeting the SHP-motif binding site of the AAA+ ATPase p97
Commun Chem, 5, 2022
|
|
8XWX
 
 | |
9EQF
 
 | | Crystal structure of the L-arginine hydroxylase VioC MeHis316, bound to Fe(II), L-arginine, and succinate | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | Authors: | Hardy, F.J. | | Deposit date: | 2024-03-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing Ferryl Reactivity in a Nonheme Iron Oxygenase Using an Expanded Genetic Code.
Acs Catalysis, 14, 2024
|
|
