4R36
 
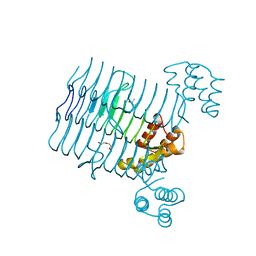 | | Crystal structure analysis of LpxA, a UDP-N-acetylglucosamine acyltransferase from Bacteroides fragilis 9343 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETATE ION, ... | | Authors: | Ngo, A, Fong, K, Cox, D, Fisher, A, Chen, X. | | Deposit date: | 2014-08-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Bacteroides fragilis uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (BfLpxA).
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2AB5
 
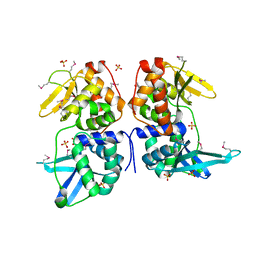 | | bI3 LAGLIDADG Maturase | | Descriptor: | SULFATE ION, mRNA maturase | | Authors: | Longo, A, Leonard, C.W, Bassi, G.S, Berndt, D, Krahn, J.M, Hall, T.M, Weeks, K.M. | | Deposit date: | 2005-07-14 | | Release date: | 2005-08-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution from DNA to RNA recognition by the bI3 LAGLIDADG maturase
Nat.Struct.Mol.Biol., 12, 2005
|
|
4JVD
 
 | |
4JVI
 
 | |
4JVC
 
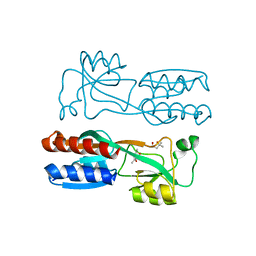 | | Crystal structure of PqsR co-inducer binding domain | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Transcriptional regulator MvfR | | Authors: | Ilangovan, A, Emsley, J, Williams, P. | | Deposit date: | 2013-03-25 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for native agonist and synthetic inhibitor recognition by the Pseudomonas aeruginosa quorum sensing regulator PqsR (MvfR).
Plos Pathog., 9, 2013
|
|
5N8O
 
 | |
2H1K
 
 | |
4R37
 
 | | Crystal structure analysis of LpxA, a UDP-N-acetylglucosamine acyltransferase from Bacteroides fragilis 9343 with UDP-GlcNAc | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETATE ION, ... | | Authors: | Fisher, A.J, Chen, X, Ngo, A. | | Deposit date: | 2014-08-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Bacteroides fragilis uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (BfLpxA).
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2QL2
 
 | | Crystal Structure of the basic-helix-loop-helix domains of the heterodimer E47/NeuroD1 bound to DNA | | Descriptor: | DNA (5'-D(*DAP*DGP*DGP*DAP*DCP*DCP*DAP*DGP*DAP*DTP*DGP*DGP*DCP*DCP*DTP*DA)-3'), DNA (5'-D(*DTP*DAP*DGP*DGP*DCP*DCP*DAP*DTP*DCP*DTP*DGP*DGP*DTP*DCP*DCP*DT)-3'), Neurogenic differentiation factor 1, ... | | Authors: | Rose, R.B, Longo, A, Guanga, G.P. | | Deposit date: | 2007-07-12 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of E47-NeuroD1/beta2 bHLH domain-DNA complex: heterodimer selectivity and DNA recognition.
Biochemistry, 47, 2008
|
|
2BPM
 
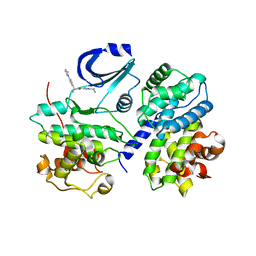 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-630529 | | Descriptor: | (2S)-N-[(3Z)-5-CYCLOPROPYL-3H-PYRAZOL-3-YLIDENE]-2-[4-(2-OXOIMIDAZOLIDIN-1-YL)PHENYL]PROPANAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Cameron, A, Fogliatto, G, Pevarello, P, Brasca, M.G, Orsini, P, Traquandi, G, Longo, A, Nesi, M, Orzi, F, Piutti, C, Sansonna, P, Varasi, M, Vulpetti, A, Roletto, F, Alzani, R, Ciomei, M, Albanese, C, Pastori, W, Marsiglio, A, Pesenti, E, Fiorentini, F, Bischoff, J.R, Mercurio, C. | | Deposit date: | 2005-04-21 | | Release date: | 2005-12-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3-Aminopyrazole Inhibitors of Cdk2-Cyclin a as Antitumor Agents. 2. Lead Optimization
J.Med.Chem., 48, 2005
|
|
6W1X
 
 | | Cryo-EM structure of anti-CRISPR AcrIF9, bound to the type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy3, ... | | Authors: | Hirschi, M, Santiago-Frangos, A, Wilkinson, R, Golden, S.M, Wiedenheft, B, Lander, G. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | AcrIF9 tethers non-sequence specific dsDNA to the CRISPR RNA-guided surveillance complex.
Nat Commun, 11, 2020
|
|
6WHI
 
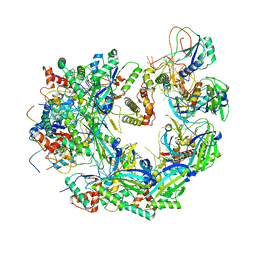 | | Cryo-electron microscopy structure of the type I-F CRISPR RNA-guided surveillance complex bound to the anti-CRISPR AcrIF9 | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy3, ... | | Authors: | Hirschi, M, Santiago-Frangos, A, Wilkinson, R, Golden, S.M, Wiedenheft, B, Lander, G. | | Deposit date: | 2020-04-08 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | AcrIF9 tethers non-sequence specific dsDNA to the CRISPR RNA-guided surveillance complex.
Nat Commun, 11, 2020
|
|
8FLJ
 
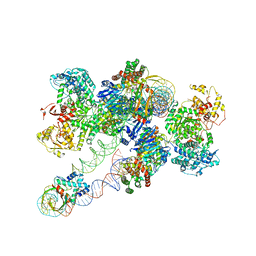 | | Cas1-Cas2/3 integrase and IHF bound to CRISPR leader, repeat and foreign DNA | | Descriptor: | CRISPR leader and repeat, anti-sense strand of DNA, CRISPR leader, ... | | Authors: | Santiago-Frangos, A, Henriques, W.S, Wiegand, T, Gauvin, C, Buyukyoruk, M, Neselu, K, Eng, E.T, Lander, G.C, Wiedenheft, B. | | Deposit date: | 2022-12-21 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure reveals why genome folding is necessary for site-specific integration of foreign DNA into CRISPR arrays.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6NE0
 
 | | Structure of double-stranded target DNA engaged Csy complex from Pseudomonas aeruginosa (PA-14) | | Descriptor: | CRISPR RNA (60-MER), CRISPR target DNA (44-MER), CRISPR-associated endonuclease Cas6/Csy4, ... | | Authors: | Chowdhury, S, Rollins, M.F, Carter, J, Golden, S.M, Miettinen, H.M, Santiago-Frangos, A, Faith, D, Lawrence, M.C, Wiedenheft, B, Lander, G.C. | | Deposit date: | 2018-12-15 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry.
Mol. Cell, 74, 2019
|
|
2WXV
 
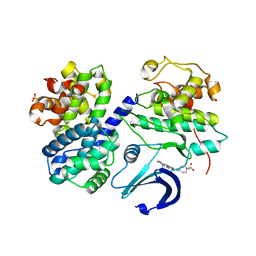 | | Structure of CDK2-CYCLIN A with a Pyrazolo(4,3-h) quinazoline-3- carboxamide inhibitor | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, N,1-DIMETHYL-8-{[1-(METHYLSULFONYL)PIPERIDIN-4-YL]AMINO}-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE, ... | | Authors: | Traquandi, G, Ciomei, M, Ballinari, D, Casale, E, Colombo, N, Croci, V, Fiorentini, F, Isacchi, A, Longo, A, Mercurio, C, Panzeri, A, Pastori, W, Pevarello, P, Volpi, D, Roussel, P, Vulpetti, A, Brasca, M.G. | | Deposit date: | 2009-11-10 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Potent Pyrazolo[4,3-H]Quinazoline-3-Carboxamides as Multi-Cyclin-Dependent Kinase Inhibitors.
J.Med.Chem., 53, 2010
|
|
7OKN
 
 | |
7OKO
 
 | |
1H56
 
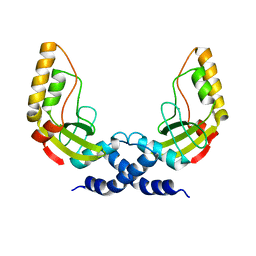 | | Structural and biochemical characterization of a new magnesium ion binding site near Tyr94 in the restriction endonuclease PvuII | | Descriptor: | MAGNESIUM ION, TYPE II RESTRICTION ENZYME PVUII | | Authors: | Spyrida, A, Matzen, C, Lanio, T, Jeltsch, A, Simoncsits, A, Athanasiadis, A, Scheuring-Vanamee, E, Kokkinidis, M, Pingoud, A. | | Deposit date: | 2001-05-20 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Biochemical Characterization of a New Mg(2+) Binding Site Near Tyr94 in the Restriction Endonuclease PvuII.
J.Mol.Biol., 331, 2003
|
|
1KTU
 
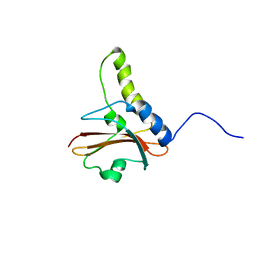 | | NuiA | | Descriptor: | NuiA | | Authors: | Kirby, T.W, Mueller, G.A, DeRose, E.F, Lebetkin, M.S, Meiss, G, Pingoud, A, London, R.E. | | Deposit date: | 2002-01-17 | | Release date: | 2002-12-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Nuclease A Inhibitor represents a new variation of the rare PR-1 fold.
J.Mol.Biol., 320, 2002
|
|
1Y42
 
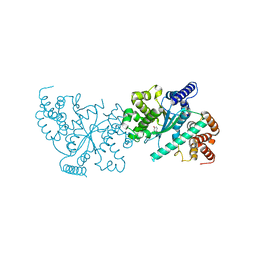 | | Crystal structure of a C-terminally truncated CYT-18 protein | | Descriptor: | TYROSINE, Tyrosyl-tRNA synthetase, mitochondrial | | Authors: | Paukstelis, P.J, Coon, R, Madabusi, L, Nowakowski, J, Monzingo, A, Robertus, J, Lambowitz, A.M. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A tyrosyl-tRNA synthetase adapted to function in group I intron splicing by acquiring a new RNA binding surface.
Mol.Cell, 17, 2005
|
|
1ZM8
 
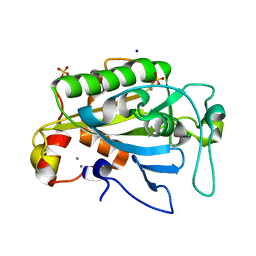 | | Apo Crystal structure of Nuclease A from Anabaena sp. | | Descriptor: | MANGANESE (II) ION, Nuclease, SODIUM ION, ... | | Authors: | Ghosh, M, Meiss, G, Pingoud, A, London, R.E, Pedersen, L.C. | | Deposit date: | 2005-05-10 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Mechanism of Nuclease A, a beta beta alpha Metal Nuclease from Anabaena.
J.Biol.Chem., 280, 2005
|
|
8OJJ
 
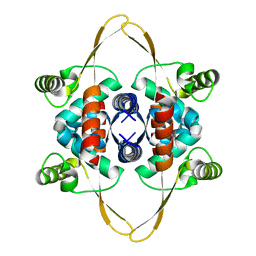 | | Cryo-EM structure of the DnaD-NTD tetramer | | Descriptor: | DNA replication protein DnaD | | Authors: | Winterhalter, C, Pelliciari, S, Cronin, N, Costa, T.R.D, Murray, H, Ilangovan, A. | | Deposit date: | 2023-03-24 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (5.47 Å) | | Cite: | The DNA replication initiation protein DnaD recognises a specific strand of the Bacillus subtilis chromosome origin.
Nucleic Acids Res., 51, 2023
|
|
7KHW
 
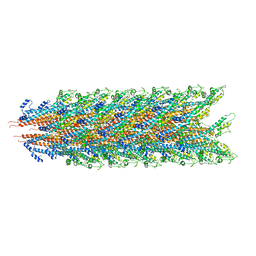 | |
4L0J
 
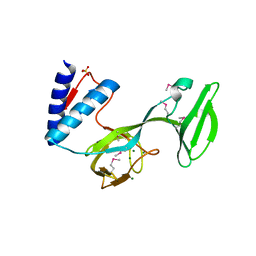 | | Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems | | Descriptor: | DNA helicase I, MAGNESIUM ION, SULFATE ION | | Authors: | Redzej, A, Ilangovan, A, Lang, S, Gruber, C.J, Topf, M, Zangger, K, Zechner, E.L, Waksman, G. | | Deposit date: | 2013-05-31 | | Release date: | 2013-06-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems.
Mol.Microbiol., 89, 2013
|
|
8BTG
 
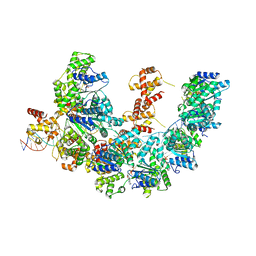 | | Cryo-EM structure of the bacterial replication origin opening basal unwinding system | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chromosomal replication initiator protein DnaA, DNA (41-MER), ... | | Authors: | Pelliciari, S, Bodet-Lefevre, S, Murray, H, Ilangovan, A. | | Deposit date: | 2022-11-28 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The bacterial replication origin BUS promotes nucleobase capture.
Nat Commun, 14, 2023
|
|
