5Y3E
 
 | |
5Y3Q
 
 | |
7CZ4
 
 | |
7C33
 
 | | Macro domain of SARS-CoV-2 in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Lin, M.H, Hsu, C.H. | | Deposit date: | 2020-05-11 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.83 Å) | | Cite: | Structural, Biophysical, and Biochemical Elucidation of the SARS-CoV-2 Nonstructural Protein 3 Macro Domain.
Acs Infect Dis., 6, 2020
|
|
7C4H
 
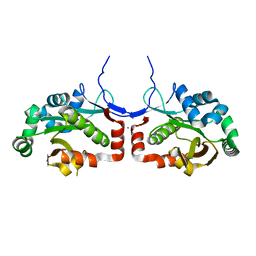 | |
7COT
 
 | |
5YB9
 
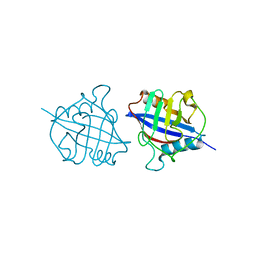 | | Crystal structure of a dimeric cyclophilin A from T.vaginalis | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Cho, C.C, Lin, M.H, Chou, C.C, Martin, T, Chen, C, Hsu, C.H. | | Deposit date: | 2017-09-04 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.276 Å) | | Cite: | Structural basis of interaction between dimeric cyclophilin 1 and Myb1 transcription factor in Trichomonas vaginalis
Sci Rep, 8, 2018
|
|
5YBA
 
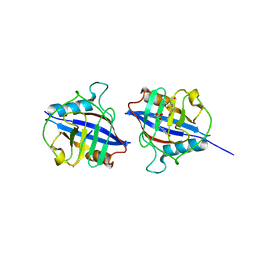 | | Dimeric Cyclophilin from T.vaginalis in complex with Myb1 peptide | | Descriptor: | Myb1 peptide, Peptidyl-prolyl cis-trans isomerase | | Authors: | Cho, C.C, Lin, M.H, Martin, T, Chou, C.C, Chen, C, Hsu, C.H. | | Deposit date: | 2017-09-04 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.062 Å) | | Cite: | Structural basis of interaction between dimeric cyclophilin 1 and Myb1 transcription factor in Trichomonas vaginalis
Sci Rep, 8, 2018
|
|
8I26
 
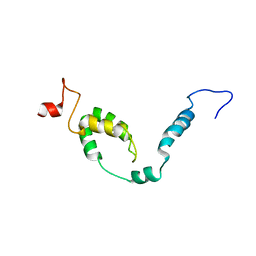 | | NMR structure of Toxoplasma gondii PDCD5 (cis form) | | Descriptor: | Programmed cell death 5 protein | | Authors: | Lin, M.H, Hsu, C.H. | | Deposit date: | 2023-01-14 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Proline Isomerization and Molten Globular Property of TgPDCD5 Secreted from Toxoplasma gondii Confers Its Regulation of Heparin Sulfate Binding.
Jacs Au, 4, 2024
|
|
8I25
 
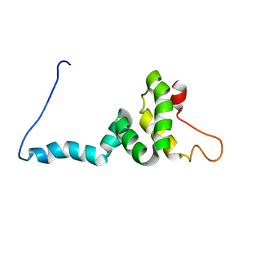 | | NMR structure of Toxoplasma gondii PDCD5 (trans form) | | Descriptor: | Programmed cell death 5 protein | | Authors: | Lin, M.H, Hsu, C.H. | | Deposit date: | 2023-01-14 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Proline Isomerization and Molten Globular Property of TgPDCD5 Secreted from Toxoplasma gondii Confers Its Regulation of Heparin Sulfate Binding.
Jacs Au, 4, 2024
|
|
8UIB
 
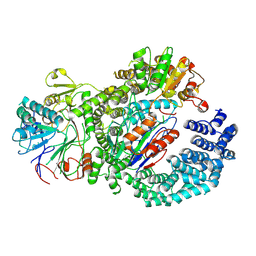 | | Structure of the human INTS9-INTS11-BRAT1 complex | | Descriptor: | BRCA1-associated ATM activator 1, Integrator complex subunit 11, Integrator complex subunit 9, ... | | Authors: | Lin, M, Tong, L. | | Deposit date: | 2023-10-10 | | Release date: | 2024-08-21 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cytoplasmic binding partners of the Integrator endonuclease INTS11 and its paralog CPSF73 are required for their nuclear function.
Mol.Cell, 84, 2024
|
|
8UIC
 
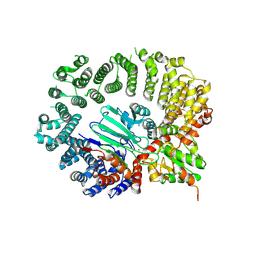 | | Structure of the Drosophila IntS11-CG7044(dBRAT1) complex | | Descriptor: | FI02071p, Integrator complex subunit 11, ZINC ION | | Authors: | Lin, M, Tong, L. | | Deposit date: | 2023-10-10 | | Release date: | 2024-08-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Cytoplasmic binding partners of the Integrator endonuclease INTS11 and its paralog CPSF73 are required for their nuclear function.
Mol.Cell, 84, 2024
|
|
7SN8
 
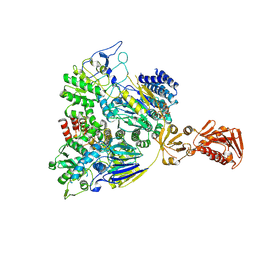 | |
4UMK
 
 | | The complex of Spo0J and parS DNA in chromosomal partition system | | Descriptor: | DNA, PROBABLE CHROMOSOME-PARTITIONING PROTEIN PARB, SULFATE ION | | Authors: | Chen, B.W, Chu, C.H, Tung, J.Y, Hsu, C.E, Hsiao, C.D, Sun, Y.J. | | Deposit date: | 2014-05-19 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.096 Å) | | Cite: | Insights into ParB spreading from the complex structure of Spo0J and parS.
Proc. Natl. Acad. Sci. U.S.A., 112, 2015
|
|
8G1U
 
 | | Structure of the methylosome-Lsm10/11 complex | | Descriptor: | ADENOSINE, Methylosome protein 50, Methylosome subunit pICln, ... | | Authors: | Lin, M, Paige, A, Tong, L. | | Deposit date: | 2023-02-03 | | Release date: | 2023-08-23 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | In vitro methylation of the U7 snRNP subunits Lsm11 and SmE by the PRMT5/MEP50/pICln methylosome.
Rna, 29, 2023
|
|
4XMO
 
 | | Crystal structure of c-Met in complex with (R)-5-(8-fluoro-3-(1-fluoro-1-(3-methoxyquinolin-6-yl)ethyl)-[1,2,4]triazolo[4,3-a]pyridin-6-yl)-3-methylisoxazole | | Descriptor: | 6-{(1R)-1-fluoro-1-[8-fluoro-6-(3-methyl-1,2-oxazol-5-yl)[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl}-3-methoxyquinoline, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Potent and Selective 8-Fluorotriazolopyridine c-Met Inhibitors.
J.Med.Chem., 58, 2015
|
|
4XYF
 
 | | Crystal structure of c-Met in complex with (S)-5-(8-fluoro-3-(1-(3-(2-methoxyethoxy)quinolin-6-yl)ethyl)-[1,2,4]triazolo[4,3-a]pyridin-6-yl)-3-methylisoxazole | | Descriptor: | 6-{(1S)-1-[8-fluoro-6-(3-methyl-1,2-oxazol-5-yl)[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl}-3-(2-methoxyethoxy)quinoline, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Potent and Selective 8-Fluorotriazolopyridine c-Met Inhibitors.
J.Med.Chem., 58, 2015
|
|
3I5N
 
 | | Crystal structure of c-Met with triazolopyridazine inhibitor 13 | | Descriptor: | 7-methoxy-N-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methyl]-1,5-naphthyridin-4-amine, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Whittington, D.A, Long, A.M, Boezio, A.A. | | Deposit date: | 2009-07-06 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and optimization of potent and selective triazolopyridazine series of c-Met inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5KS8
 
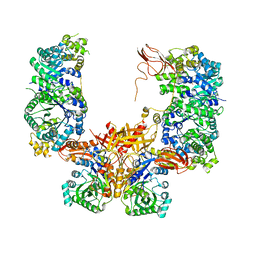 | |
5B7C
 
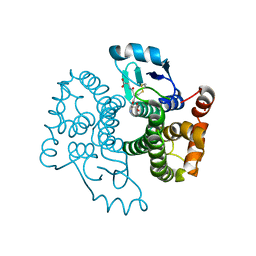 | |
5EYC
 
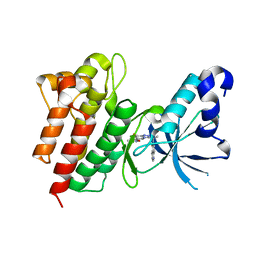 | | Crystal structure of c-Met in complex with naphthyridinone inhibitor 5 | | Descriptor: | 6-[(1~{R})-1-[8-fluoranyl-6-(3-methyl-1,2-oxazol-5-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl]-1,6-naphthyridin-5-one, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of (R)-6-(1-(8-Fluoro-6-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl)ethyl)-3-(2-methoxyethoxy)-1,6-naphthyridin-5(6H)-one (AMG 337), a Potent and Selective Inhibitor of MET with High Unbound Target Coverage and Robust In Vivo Antitumor Activity.
J.Med.Chem., 59, 2016
|
|
5EYD
 
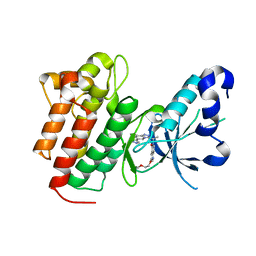 | | Crystal structure of c-Met in complex with AMG 337 | | Descriptor: | 6-[(1~{R})-1-[8-fluoranyl-6-(1-methylpyrazol-4-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl]-3-(2-methoxyethoxy)-1,6-naphthyridin-5-one, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of (R)-6-(1-(8-Fluoro-6-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl)ethyl)-3-(2-methoxyethoxy)-1,6-naphthyridin-5(6H)-one (AMG 337), a Potent and Selective Inhibitor of MET with High Unbound Target Coverage and Robust In Vivo Antitumor Activity.
J.Med.Chem., 59, 2016
|
|
7Y0G
 
 | | Crystal structure of anti-mPEG h15-2b Fab | | Descriptor: | 15-2b heavy chain, 15-2b light chain, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL | | Authors: | Chang, C.Y, Nguyen, T.M.T, Lin, E.C, Su, Y.C. | | Deposit date: | 2022-06-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural determination of an antibody that specifically recognizes polyethylene glycol with a terminal methoxy group.
Commun Chem, 5, 2022
|
|
7YIZ
 
 | | Crystal structure of anti-mPEG h15-2b Fab W104Y mutant | | Descriptor: | 15-2b W104Y heavy chain, 15-2b light chain, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL | | Authors: | Chang, C.Y, Nguyen, T.M.T, Li, Y.C, Su, Y.C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-08-02 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Engineering a High-Affinity Anti-Methoxy Poly(ethylene glycol) (mPEG) Antibody for Sensitive Immunosensing of mPEGylated Therapeutics and mPEG Molecules
Bioconjug.Chem., 33, 2022
|
|
8Z95
 
 | | Humanized anti-PEG h6.3 Fab in complex with PEG | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, Heavy chain of Anti-PEG antibody h6-3 Fab fragment, ISOPROPYL ALCOHOL, ... | | Authors: | Lin, Y.C, Chang, C.Y, Su, Y.C. | | Deposit date: | 2024-04-22 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | pH-Responsive Polyethylene Glycol Engagers for Enhanced Brain Delivery of PEGylated Nanomedicine to Treat Glioblastoma.
Acs Nano, 19, 2025
|
|
