2L8P
 
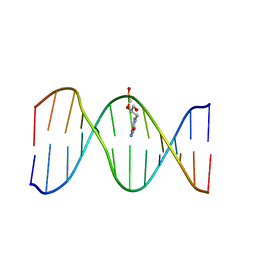 | | Solution Structure of a DNA Duplex Containing the Potent Anti-Poxvirus Agent Cidofovir | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*TP*GP*(L8P)P*TP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*AP*GP*CP*AP*TP*GP*CP*G)-3') | | Authors: | Julien, O, Beadle, J.R, Magee, W.C, Chatterjee, S, Hostetler, K.Y, Evans, D.H, Sykes, B.D. | | Deposit date: | 2011-01-22 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA duplex containing the potent anti-poxvirus agent cidofovir.
J.Am.Chem.Soc., 133, 2011
|
|
2JXL
 
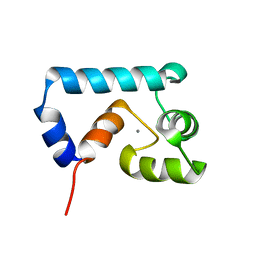 | | Solution structure of cardiac N-domain troponin C mutant F77W-V82A | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles | | Authors: | Julien, O, Sun, Y, Wang, X, Lindhout, D.A, Thiessen, A, Irving, M, Sykes, B.D. | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Tryptophan Mutants of Cardiac Troponin C: 3D Structure, Troponin I Affinity, and in Situ Activity.
Biochemistry, 47, 2008
|
|
2L8Q
 
 | | Solution Structure of a control DNA Duplex | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*TP*GP*CP*TP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*AP*GP*CP*AP*TP*GP*CP*G)-3') | | Authors: | Julien, O, Beadle, J.R, Magee, W.C, Chatterjee, S, Hostetler, K.Y, Evans, D.H, Sykes, B.D. | | Deposit date: | 2011-01-22 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA duplex containing the potent anti-poxvirus agent cidofovir.
J.Am.Chem.Soc., 133, 2011
|
|
1TE4
 
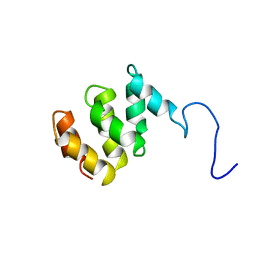 | | Solution structure of MTH187. Ontario Centre for Structural Proteomics target MTH0187_1_111; Northeast Structural Genomics Target TT740 | | Descriptor: | conserved protein MTH187 | | Authors: | Gignac, I, Julien, O, Yee, A, Arrowsmith, C.H, Gagne, S.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-05-24 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | MTH187 from Methanobacterium thermoautotrophicum has three HEAT-like Repeats.
J.Biomol.Nmr, 35, 2006
|
|
2KGB
 
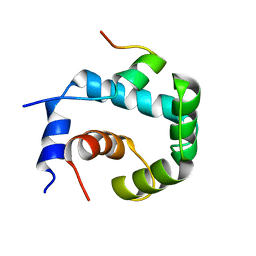 | | NMR solution of the regulatory domain cardiac F77W-Troponin C in complex with the cardiac Troponin I 144-163 switch peptide | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles, ... | | Authors: | Mercier, P, Julien, O, Crane, M.L, Sykes, B.D. | | Deposit date: | 2009-03-07 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The effect of the cosolvent trifluoroethanol on a tryptophan side chain orientation in the hydrophobic core of troponin C.
Protein Sci., 18, 2009
|
|
5IC6
 
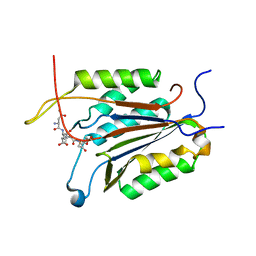 | | Crystal structure of caspase-7 DEVE peptide complex | | Descriptor: | Caspase-7 subunit p11, Caspase-7 subunit p20, DEVE peptide | | Authors: | Seaman, J.E, Julien, O, Lee, P.S, Rettenmaier, T.J, Thomsen, N.D, Wells, J.A. | | Deposit date: | 2016-02-22 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cacidases: caspases can cleave after aspartate, glutamate and phosphoserine residues.
Cell Death Differ., 23, 2016
|
|
5IC4
 
 | | Crystal structure of caspase-3 DEVE peptide complex | | Descriptor: | Caspase-3 subunit p12, Caspase-3 subunit p17, DEVE peptide | | Authors: | Seaman, J.E, Julien, O, Lee, P.S, Rettenmaier, T.J, Thomsen, N.D, Wells, J.A. | | Deposit date: | 2016-02-22 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Cacidases: caspases can cleave after aspartate, glutamate and phosphoserine residues.
Cell Death Differ., 23, 2016
|
|
4ZVR
 
 | |
4ZVT
 
 | |
4ZVP
 
 | |
4ZVU
 
 | |
4ZVQ
 
 | |
4ZVS
 
 | |
4ZVO
 
 | |
8DKZ
 
 | |
8DJJ
 
 | |
8DI3
 
 | |
8DKK
 
 | |
8DK8
 
 | |
8DKH
 
 | |
8DKJ
 
 | |
8DKL
 
 | |
8DMN
 
 | |
8EJ9
 
 | |
8EJ7
 
 | |
